Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
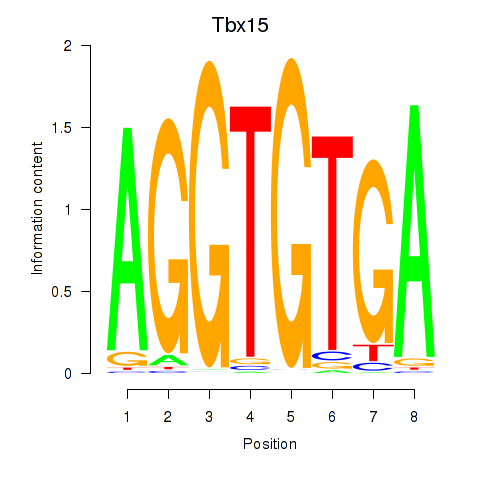
Results for Tbx15
Z-value: 0.95
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSMUSG00000027868.12 | T-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | mm39_v1_chr3_+_99161070_99161122 | -0.19 | 2.7e-01 | Click! |
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20579322 | 8.69 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr7_+_25597045 | 6.34 |
ENSMUST00000072438.13
ENSMUST00000005477.6 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chr7_-_100307601 | 5.09 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_+_87697155 | 4.85 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr19_+_37686240 | 3.53 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr7_-_100307571 | 3.43 |
ENSMUST00000107043.8
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr11_-_74816484 | 3.23 |
ENSMUST00000138612.2
ENSMUST00000123855.8 ENSMUST00000128556.8 ENSMUST00000108448.8 ENSMUST00000108447.8 ENSMUST00000065211.9 |
Srr
|
serine racemase |
| chr3_-_129126362 | 2.89 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr10_-_86541349 | 2.88 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr11_-_74816750 | 2.81 |
ENSMUST00000121738.8
|
Srr
|
serine racemase |
| chr18_-_75094323 | 2.62 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr4_-_129132963 | 2.61 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr9_-_43151179 | 2.54 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr6_-_124894902 | 2.53 |
ENSMUST00000032216.7
|
Ptms
|
parathymosin |
| chr16_-_18232202 | 2.53 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr2_+_58645189 | 2.53 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr1_-_65225617 | 2.49 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr2_+_58644922 | 2.40 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr4_-_107975723 | 2.38 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr2_+_164404499 | 2.38 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr16_-_23807602 | 2.37 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr15_-_60793115 | 2.11 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr4_+_43641262 | 2.10 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr2_+_26969384 | 2.10 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr1_-_65225572 | 2.09 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_+_43655230 | 2.06 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr11_+_95304903 | 1.99 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr3_-_107851021 | 1.97 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_-_35081129 | 1.97 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr6_+_125297596 | 1.88 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr9_+_47441471 | 1.85 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr7_+_44034225 | 1.83 |
ENSMUST00000120262.2
|
Syt3
|
synaptotagmin III |
| chr15_+_57558048 | 1.83 |
ENSMUST00000096430.11
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr4_-_129121676 | 1.81 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr19_+_37425180 | 1.75 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr2_-_147887810 | 1.70 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr11_-_72686627 | 1.70 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr14_-_30665232 | 1.69 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr9_+_108174052 | 1.69 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr19_-_43512929 | 1.66 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chrX_+_138464065 | 1.65 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr1_-_105284383 | 1.65 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr4_+_144619647 | 1.62 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr15_-_81244940 | 1.62 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr3_-_75464066 | 1.60 |
ENSMUST00000162138.2
ENSMUST00000029424.12 ENSMUST00000161137.8 |
Pdcd10
|
programmed cell death 10 |
| chr19_-_42190589 | 1.59 |
ENSMUST00000018966.8
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr3_+_63203516 | 1.58 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr3_-_89294430 | 1.56 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr2_+_71811526 | 1.54 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr10_-_10433831 | 1.54 |
ENSMUST00000019974.5
|
Rab32
|
RAB32, member RAS oncogene family |
| chr4_+_53440516 | 1.54 |
ENSMUST00000107651.9
ENSMUST00000107647.8 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr9_+_102885156 | 1.53 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr1_-_72323464 | 1.51 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_-_72323407 | 1.47 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr16_+_31482745 | 1.46 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_118692435 | 1.46 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr7_-_126497421 | 1.44 |
ENSMUST00000121532.8
ENSMUST00000032926.12 |
Tmem219
|
transmembrane protein 219 |
| chr4_+_84802592 | 1.42 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr18_+_36414122 | 1.41 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chrX_-_51254129 | 1.41 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr2_+_30254239 | 1.40 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr17_-_74354844 | 1.39 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chrX_+_100427331 | 1.36 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr1_-_105284407 | 1.33 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr16_+_31482658 | 1.33 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr6_+_108805594 | 1.28 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chrX_-_138683102 | 1.26 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr7_-_28078671 | 1.26 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr7_-_34353767 | 1.25 |
ENSMUST00000206501.2
ENSMUST00000108069.8 |
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr19_-_37340010 | 1.24 |
ENSMUST00000131070.3
|
Ide
|
insulin degrading enzyme |
| chr16_+_31482949 | 1.24 |
ENSMUST00000023454.12
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr4_+_84802513 | 1.23 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr2_-_180844582 | 1.23 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr2_+_32498997 | 1.20 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr4_-_133066549 | 1.20 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr15_+_31565508 | 1.20 |
ENSMUST00000226951.2
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr10_+_86541675 | 1.18 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr2_+_143757193 | 1.17 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr4_-_133066594 | 1.17 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chrM_+_10167 | 1.16 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_+_180720666 | 1.15 |
ENSMUST00000085797.6
|
Lefty2
|
left-right determination factor 2 |
| chr7_-_28001624 | 1.13 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chrX_+_150799414 | 1.13 |
ENSMUST00000045312.6
|
Smc1a
|
structural maintenance of chromosomes 1A |
| chr5_+_120651158 | 1.10 |
ENSMUST00000111889.2
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr11_-_23720953 | 1.08 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr3_+_59939175 | 1.07 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr16_+_22926162 | 1.07 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr11_-_51748450 | 1.07 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr15_-_10714653 | 1.05 |
ENSMUST00000169385.3
|
Rai14
|
retinoic acid induced 14 |
| chr3_+_40905066 | 1.05 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr3_+_122277372 | 1.05 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_-_50657800 | 1.03 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chrX_+_41239872 | 1.02 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr14_-_45626237 | 1.02 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chrM_+_9459 | 1.01 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_-_42168603 | 1.01 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 1.00 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr3_-_115508680 | 1.00 |
ENSMUST00000055676.4
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr3_+_121517158 | 1.00 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chr12_-_35584968 | 0.98 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr3_-_121056944 | 0.98 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr10_-_34294461 | 0.97 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr11_+_78085522 | 0.97 |
ENSMUST00000060539.13
|
Proca1
|
protein interacting with cyclin A1 |
| chrM_+_9870 | 0.96 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_153541020 | 0.96 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr5_-_147244074 | 0.96 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr16_-_9812410 | 0.95 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr6_+_108190163 | 0.95 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr4_+_48045143 | 0.95 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_+_121798932 | 0.95 |
ENSMUST00000111765.8
|
Brap
|
BRCA1 associated protein |
| chr14_-_55880708 | 0.95 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr1_+_106099482 | 0.94 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr15_-_82783978 | 0.94 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr13_+_64309675 | 0.93 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr18_+_36693024 | 0.93 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr6_+_121320339 | 0.92 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr4_+_155648256 | 0.92 |
ENSMUST00000143840.2
ENSMUST00000146080.8 |
Nadk
|
NAD kinase |
| chr16_-_56987670 | 0.91 |
ENSMUST00000023432.10
|
Nit2
|
nitrilase family, member 2 |
| chr11_-_98040377 | 0.91 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr7_+_127070615 | 0.89 |
ENSMUST00000033095.10
|
Prr14
|
proline rich 14 |
| chr4_+_84802650 | 0.88 |
ENSMUST00000169371.9
|
Cntln
|
centlein, centrosomal protein |
| chr3_-_142101418 | 0.88 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr14_+_53995108 | 0.88 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr3_+_75464837 | 0.88 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr1_-_40829801 | 0.87 |
ENSMUST00000039672.6
|
Mfsd9
|
major facilitator superfamily domain containing 9 |
| chr2_+_28403084 | 0.87 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr2_+_140237366 | 0.86 |
ENSMUST00000110061.2
|
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr7_+_101027390 | 0.86 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_+_78085560 | 0.86 |
ENSMUST00000108317.3
|
Proca1
|
protein interacting with cyclin A1 |
| chr7_+_27770655 | 0.86 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr3_+_40905216 | 0.85 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr9_-_22042930 | 0.85 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr16_+_62635039 | 0.84 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr6_+_124281607 | 0.84 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chr2_-_147888816 | 0.84 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr9_-_20657643 | 0.84 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr8_+_111760521 | 0.84 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr18_-_46345661 | 0.83 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr12_+_10440755 | 0.83 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr15_-_44291226 | 0.83 |
ENSMUST00000227843.2
|
Nudcd1
|
NudC domain containing 1 |
| chr7_-_126496534 | 0.82 |
ENSMUST00000120007.8
|
Tmem219
|
transmembrane protein 219 |
| chr2_-_101459274 | 0.82 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr7_-_4607040 | 0.80 |
ENSMUST00000166650.3
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr4_-_82423985 | 0.80 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr2_-_101459303 | 0.80 |
ENSMUST00000111231.10
|
Iftap
|
intraflagellar transport associated protein |
| chr4_-_63779562 | 0.80 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr6_+_50087167 | 0.80 |
ENSMUST00000166318.8
ENSMUST00000036236.15 ENSMUST00000204545.3 ENSMUST00000036225.15 |
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr14_+_58313964 | 0.79 |
ENSMUST00000166770.2
|
Fgf9
|
fibroblast growth factor 9 |
| chr9_-_72946980 | 0.79 |
ENSMUST00000184035.8
ENSMUST00000098566.5 |
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr9_-_121745354 | 0.79 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr2_-_172782089 | 0.79 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr15_-_100583044 | 0.78 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr13_-_111626562 | 0.78 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr5_+_121798623 | 0.78 |
ENSMUST00000031414.15
|
Brap
|
BRCA1 associated protein |
| chr7_-_78228116 | 0.77 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr2_-_38816229 | 0.77 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr11_-_86698484 | 0.77 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr2_+_140237229 | 0.76 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr11_+_68858942 | 0.76 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr1_-_57010921 | 0.76 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr7_-_4606104 | 0.75 |
ENSMUST00000049113.14
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr1_-_63215952 | 0.75 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_+_163662752 | 0.75 |
ENSMUST00000029188.8
|
Ccn5
|
cellular communication network factor 5 |
| chrX_+_167819606 | 0.74 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr7_-_119694400 | 0.73 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr17_+_44114894 | 0.72 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr7_+_4240697 | 0.72 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr16_+_48104098 | 0.71 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
| chr9_-_58109627 | 0.71 |
ENSMUST00000216231.2
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr6_+_42263644 | 0.71 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr9_-_57347366 | 0.70 |
ENSMUST00000214144.2
ENSMUST00000085709.6 ENSMUST00000214624.2 ENSMUST00000215883.2 ENSMUST00000214339.2 ENSMUST00000215299.2 ENSMUST00000214166.2 ENSMUST00000214065.2 |
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr7_+_44034028 | 0.70 |
ENSMUST00000130707.8
ENSMUST00000130844.3 |
Syt3
|
synaptotagmin III |
| chr2_+_136733421 | 0.70 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr19_+_21249636 | 0.70 |
ENSMUST00000237651.2
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr2_+_154975419 | 0.70 |
ENSMUST00000029126.15
ENSMUST00000109685.8 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr14_+_53399856 | 0.70 |
ENSMUST00000198359.2
|
Trav13n-1
|
T cell receptor alpha variable 13N-1 |
| chr6_+_42263609 | 0.70 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr4_-_130172998 | 0.70 |
ENSMUST00000120126.9
|
Serinc2
|
serine incorporator 2 |
| chr11_-_86574586 | 0.70 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr9_-_22043083 | 0.69 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr5_+_143450329 | 0.69 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr4_+_148085179 | 0.69 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr7_+_107194446 | 0.69 |
ENSMUST00000040056.15
ENSMUST00000208956.2 |
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr6_+_108190050 | 0.67 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr16_+_84631956 | 0.67 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr8_+_95328694 | 0.66 |
ENSMUST00000211983.2
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr17_-_31363245 | 0.66 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr6_+_7555053 | 0.66 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr4_-_57300361 | 0.66 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr4_-_41517326 | 0.66 |
ENSMUST00000030152.13
ENSMUST00000095126.5 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr6_+_115111872 | 0.66 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr9_-_72946928 | 0.66 |
ENSMUST00000184389.8
|
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr3_-_84128160 | 0.65 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr10_-_120815232 | 0.65 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr15_-_76004395 | 0.65 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr7_+_105289729 | 0.64 |
ENSMUST00000210350.2
ENSMUST00000151193.2 ENSMUST00000209588.2 ENSMUST00000106780.2 ENSMUST00000106784.2 ENSMUST00000106785.8 ENSMUST00000106786.8 ENSMUST00000211054.2 |
Timm10b
Gm45799
|
translocase of inner mitochondrial membrane 10B predicted gene 45799 |
| chr9_+_72946994 | 0.64 |
ENSMUST00000184126.3
|
Pigbos1
|
Pigb opposite strand 1 |
| chr12_+_33364288 | 0.64 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr6_-_28261881 | 0.63 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr14_+_53698556 | 0.63 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 1.1 | 4.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.0 | 4.0 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 1.0 | 3.0 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.9 | 6.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.9 | 8.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 2.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.7 | 4.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 2.6 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.6 | 2.5 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.6 | 2.4 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.6 | 2.4 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.6 | 1.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 1.7 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.5 | 1.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.5 | 1.6 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 3.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.4 | 1.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.4 | 1.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 2.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.4 | 1.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 1.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 1.9 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 1.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 1.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.4 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 1.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 1.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 2.0 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 1.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 1.7 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.3 | 1.7 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.3 | 2.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 2.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 1.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 1.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 1.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 2.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 1.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.9 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 0.7 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.2 | 1.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.7 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.2 | 1.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 1.5 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.6 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 | 0.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 1.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.6 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 0.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 0.9 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.2 | 1.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 2.9 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.5 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 1.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 2.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 1.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 1.5 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.2 | 0.5 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:0090327 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 2.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 1.7 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 2.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 1.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.6 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 0.6 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.4 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.6 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 0.2 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 2.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 1.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.6 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 2.1 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 2.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.0 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.7 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 1.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 1.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.2 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.1 | 0.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 0.5 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 1.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.6 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.8 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.6 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.5 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.1 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.1 | 0.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.4 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.4 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 1.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.4 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 0.3 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.8 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:1904346 | gastric emptying(GO:0035483) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 1.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.6 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) photoreceptor cell outer segment organization(GO:0035845) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.3 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 2.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 1.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 2.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.5 | GO:0002664 | regulation of T cell tolerance induction(GO:0002664) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 1.9 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.5 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.7 | GO:0090208 | positive regulation of triglyceride biosynthetic process(GO:0010867) positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.0 | 1.0 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.9 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 2.3 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 4.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0051771 | protein farnesylation(GO:0018343) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0032324 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.9 | GO:0001662 | behavioral fear response(GO:0001662) fear response(GO:0042596) |
| 0.0 | 0.2 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.4 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.0 | GO:1900175 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 1.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0090500 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.1 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.8 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 4.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 4.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 1.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.4 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 1.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 0.9 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.3 | 2.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 1.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 0.9 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.2 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 2.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 3.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.6 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 1.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 8.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 2.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 3.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 2.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 7.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 1.7 | 8.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.1 | 4.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.8 | 2.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.8 | 4.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.7 | 3.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.6 | 2.4 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.6 | 2.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.6 | 3.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.6 | 4.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.6 | 1.7 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.5 | 2.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 1.5 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.4 | 2.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 4.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 1.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 1.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 1.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 1.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 1.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 1.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.2 | 1.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.0 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 6.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 2.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 3.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.4 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 2.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 4.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.8 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.6 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.0 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 1.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0050693 | DBD domain binding(GO:0050692) LBD domain binding(GO:0050693) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 8.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.5 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.1 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 4.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.0 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 6.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 3.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 3.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 8.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 4.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |