Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
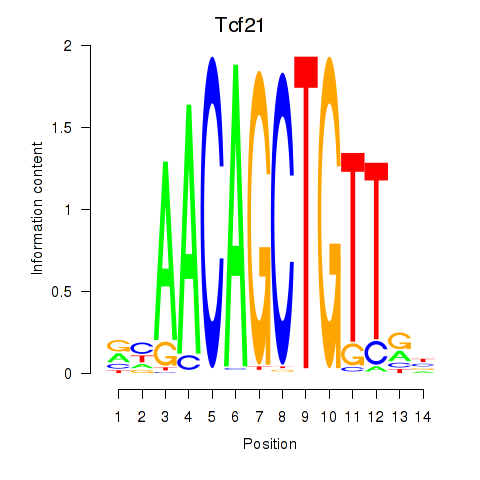
Results for Tcf21_Msc
Z-value: 0.96
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf21
|
ENSMUSG00000045680.9 | transcription factor 21 |
|
Msc
|
ENSMUSG00000025930.7 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msc | mm39_v1_chr1_-_14826208_14826222 | 0.49 | 2.4e-03 | Click! |
| Tcf21 | mm39_v1_chr10_-_22696025_22696073 | 0.38 | 2.3e-02 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_100922910 | 8.19 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr11_-_102255999 | 7.46 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr5_+_123214332 | 7.42 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr4_-_87724533 | 7.02 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_37180437 | 6.14 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr17_-_26417982 | 5.87 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr5_-_145406533 | 5.30 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr7_-_142223662 | 5.16 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr8_-_123159663 | 5.11 |
ENSMUST00000017604.10
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr8_-_123159639 | 4.93 |
ENSMUST00000212600.2
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr2_-_122441719 | 4.76 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_+_95496270 | 4.50 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr11_+_104468107 | 4.49 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_+_29694181 | 4.45 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr12_+_109425769 | 4.43 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr9_-_114610879 | 4.40 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr7_+_44866635 | 4.39 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr7_-_126303947 | 4.37 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_+_95496239 | 4.31 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr12_+_24758240 | 4.26 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr7_+_142558783 | 4.25 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr4_-_87724512 | 4.16 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr19_-_15901919 | 3.76 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_-_100396635 | 3.69 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr7_+_142558837 | 3.56 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr11_+_104467791 | 3.43 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr7_+_97492124 | 3.26 |
ENSMUST00000033040.12
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr7_-_127593003 | 3.15 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr5_-_138169509 | 2.97 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr2_+_130116357 | 2.96 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr2_-_120867529 | 2.91 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr2_+_157401998 | 2.88 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr5_-_145742672 | 2.87 |
ENSMUST00000067479.6
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr15_+_78810919 | 2.82 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr11_+_99748741 | 2.80 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr13_+_95833359 | 2.76 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr11_-_5753693 | 2.74 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr3_-_129625023 | 2.68 |
ENSMUST00000029643.15
|
Gar1
|
GAR1 ribonucleoprotein |
| chr17_-_7652863 | 2.67 |
ENSMUST00000070059.5
|
Unc93a2
|
unc-93 homolog A2 |
| chr5_-_145656934 | 2.63 |
ENSMUST00000094111.6
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr6_+_145091196 | 2.59 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr17_-_34170401 | 2.57 |
ENSMUST00000087543.5
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr7_-_126303887 | 2.52 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr5_-_145521533 | 2.52 |
ENSMUST00000075837.8
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr9_+_118892497 | 2.52 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr14_-_20319242 | 2.48 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr1_+_130659700 | 2.45 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr2_+_84564394 | 2.44 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr12_+_24758724 | 2.27 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_+_103148824 | 2.22 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr17_-_35304582 | 2.19 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr6_-_121058551 | 2.17 |
ENSMUST00000077159.8
ENSMUST00000207889.2 ENSMUST00000238957.2 ENSMUST00000238920.2 ENSMUST00000238968.2 |
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr7_-_45173193 | 2.13 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr5_+_35146727 | 2.13 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_-_182929025 | 2.13 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr4_-_116228921 | 2.11 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_84670956 | 2.10 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr8_+_95703728 | 2.10 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_40624200 | 2.10 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr16_+_17269845 | 2.07 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr11_+_115790768 | 2.06 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr16_+_45430743 | 2.03 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr5_-_138170077 | 2.01 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr11_+_117877118 | 1.99 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr15_-_100567377 | 1.96 |
ENSMUST00000182814.8
ENSMUST00000238935.2 ENSMUST00000182068.8 ENSMUST00000182574.2 ENSMUST00000182775.8 |
Bin2
|
bridging integrator 2 |
| chr4_-_133599616 | 1.96 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr7_+_110371811 | 1.93 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr17_-_43813664 | 1.92 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr4_-_133600308 | 1.90 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr13_-_97897139 | 1.90 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr10_+_82821304 | 1.89 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr17_+_47983587 | 1.88 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr4_-_131695135 | 1.87 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chrX_+_74557905 | 1.84 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr8_-_108151661 | 1.84 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr10_+_127871444 | 1.81 |
ENSMUST00000073868.9
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr4_-_137157824 | 1.80 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr17_+_56259617 | 1.79 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr13_-_33035150 | 1.77 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr4_-_156339769 | 1.76 |
ENSMUST00000217934.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr7_-_126303689 | 1.74 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_+_142025817 | 1.73 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr3_+_96465265 | 1.73 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr11_+_115790951 | 1.70 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr17_-_7639378 | 1.68 |
ENSMUST00000231397.2
|
Gm49630
|
predicted gene, 49630 |
| chr13_-_74210360 | 1.66 |
ENSMUST00000222609.2
ENSMUST00000036456.8 |
Cep72
|
centrosomal protein 72 |
| chr7_+_142025575 | 1.66 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr1_-_172722589 | 1.63 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr1_-_133352115 | 1.61 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr10_-_7539009 | 1.60 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr7_+_28488380 | 1.59 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr10_-_62343516 | 1.58 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr10_-_88192852 | 1.56 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_-_113968483 | 1.56 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr5_+_103902426 | 1.54 |
ENSMUST00000153165.8
ENSMUST00000031256.6 |
Aff1
|
AF4/FMR2 family, member 1 |
| chr6_-_57827328 | 1.53 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr4_-_133694607 | 1.53 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_+_5118103 | 1.52 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr10_+_127871493 | 1.52 |
ENSMUST00000092048.13
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr17_+_28059129 | 1.51 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr8_-_25592001 | 1.50 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr7_-_141579689 | 1.49 |
ENSMUST00000209732.2
|
Mob2
|
MOB kinase activator 2 |
| chr7_+_28140352 | 1.49 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr11_+_61017573 | 1.49 |
ENSMUST00000010286.8
ENSMUST00000146033.8 ENSMUST00000139422.8 |
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b |
| chr11_+_32155415 | 1.48 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chrX_+_55500170 | 1.48 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr1_+_180762587 | 1.47 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr11_-_79971750 | 1.47 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr5_-_138169476 | 1.45 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_+_31329939 | 1.44 |
ENSMUST00000236241.2
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr4_-_133695264 | 1.43 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr16_+_21828515 | 1.43 |
ENSMUST00000231632.2
ENSMUST00000232534.2 |
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr2_+_26863428 | 1.40 |
ENSMUST00000014996.14
|
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
| chr11_+_101207743 | 1.39 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr3_-_105708601 | 1.38 |
ENSMUST00000197094.5
ENSMUST00000198004.2 |
Rap1a
|
RAS-related protein 1a |
| chr14_+_13803477 | 1.37 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing |
| chr16_+_49620883 | 1.37 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr1_-_88133472 | 1.36 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr11_+_32155483 | 1.35 |
ENSMUST00000121182.2
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr17_+_28059099 | 1.34 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr13_-_55676334 | 1.34 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr19_+_11493825 | 1.33 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr3_-_81981946 | 1.33 |
ENSMUST00000029635.14
ENSMUST00000193597.2 |
Gucy1b1
|
guanylate cyclase 1, soluble, beta 1 |
| chr10_+_60182630 | 1.32 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr8_+_122317100 | 1.31 |
ENSMUST00000034270.17
ENSMUST00000181826.2 ENSMUST00000181948.2 |
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr8_+_23629046 | 1.30 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr3_+_107137924 | 1.26 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr9_+_110856425 | 1.26 |
ENSMUST00000199313.2
|
Ltf
|
lactotransferrin |
| chr7_-_44888532 | 1.25 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr17_+_75772475 | 1.24 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr18_+_50184769 | 1.23 |
ENSMUST00000134348.8
ENSMUST00000153873.3 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_4505493 | 1.23 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr1_-_192880260 | 1.22 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr12_-_86125793 | 1.21 |
ENSMUST00000003687.8
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr17_+_28059036 | 1.20 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr17_+_23945310 | 1.20 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr16_-_19801781 | 1.20 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr15_-_100567412 | 1.20 |
ENSMUST00000183211.8
|
Bin2
|
bridging integrator 2 |
| chr7_+_28140450 | 1.16 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr18_-_62313019 | 1.15 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr5_-_73805063 | 1.15 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr9_-_40257586 | 1.14 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr11_-_3321307 | 1.14 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr13_-_113182891 | 1.14 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr11_+_72580823 | 1.14 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr17_-_26420300 | 1.13 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr17_-_24424456 | 1.12 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr2_-_92222979 | 1.11 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chrX_+_100492684 | 1.11 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr5_+_123887759 | 1.10 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr12_-_40087393 | 1.10 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_+_80507671 | 1.10 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chrX_-_101295787 | 1.09 |
ENSMUST00000050551.10
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr2_-_169973076 | 1.09 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr11_+_106642052 | 1.08 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr16_+_57173456 | 1.07 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr2_-_35322581 | 1.07 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr19_-_3979723 | 1.07 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr11_+_63023893 | 1.07 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr7_+_44866095 | 1.06 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr7_+_3339077 | 1.05 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_-_107607343 | 1.05 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr2_-_73316053 | 1.05 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr14_-_54891073 | 1.04 |
ENSMUST00000126166.8
ENSMUST00000141453.8 ENSMUST00000150371.8 ENSMUST00000123875.2 ENSMUST00000022794.14 ENSMUST00000148754.10 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_+_128069677 | 1.04 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr16_+_17437205 | 1.03 |
ENSMUST00000006053.13
ENSMUST00000231311.2 ENSMUST00000163476.10 ENSMUST00000231257.2 ENSMUST00000165363.10 ENSMUST00000232043.2 ENSMUST00000090159.13 ENSMUST00000232442.2 |
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr1_+_88154727 | 1.03 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr7_+_3339059 | 1.03 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_+_51858476 | 1.03 |
ENSMUST00000102763.5
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr1_+_135727228 | 1.03 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr18_+_89215575 | 1.03 |
ENSMUST00000097496.4
ENSMUST00000236452.2 |
Cd226
|
CD226 antigen |
| chr3_+_87813624 | 1.02 |
ENSMUST00000005017.15
|
Hdgf
|
heparin binding growth factor |
| chr5_+_137639538 | 1.02 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr2_-_170269748 | 1.02 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_-_7539333 | 1.00 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr15_+_102058936 | 0.99 |
ENSMUST00000023806.14
|
Soat2
|
sterol O-acyltransferase 2 |
| chr13_-_55677109 | 0.98 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr11_+_4959515 | 0.98 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr6_-_29212295 | 0.98 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr13_+_55468313 | 0.97 |
ENSMUST00000021942.8
|
Prelid1
|
PRELI domain containing 1 |
| chr2_+_124910037 | 0.97 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr5_+_25452342 | 0.95 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr11_-_100036792 | 0.95 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr17_-_26420332 | 0.94 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr6_-_29212239 | 0.94 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr3_+_107784543 | 0.93 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr9_-_66950991 | 0.93 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr15_-_77854711 | 0.91 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr4_+_3678108 | 0.90 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr7_-_115637970 | 0.90 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_55310724 | 0.89 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr4_-_148529187 | 0.89 |
ENSMUST00000051633.3
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr5_-_123663440 | 0.89 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr17_+_71923210 | 0.88 |
ENSMUST00000047086.10
|
Wdr43
|
WD repeat domain 43 |
| chr7_-_140697719 | 0.88 |
ENSMUST00000067836.9
|
Ano9
|
anoctamin 9 |
| chr2_+_32253692 | 0.86 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr14_+_61547202 | 0.86 |
ENSMUST00000055159.8
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr2_-_92201342 | 0.85 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr7_-_126046814 | 0.85 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr5_+_64317550 | 0.84 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 2.0 | 8.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.6 | 4.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.1 | 4.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.1 | 8.6 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.1 | 3.2 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.9 | 2.8 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.9 | 2.6 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.8 | 3.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.8 | 8.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.7 | 2.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.7 | 2.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.6 | 3.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.6 | 4.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.6 | 9.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.6 | 11.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.5 | 1.6 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.5 | 2.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.5 | 6.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 1.8 | GO:0033367 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.5 | 2.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 4.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 3.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 1.7 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.4 | 2.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 | 1.7 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.4 | 1.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.4 | 3.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 1.9 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 1.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.4 | 2.5 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 | 1.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.3 | 1.3 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 1.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.3 | 1.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 0.9 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.3 | 1.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 0.9 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 1.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.3 | 6.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 7.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 1.1 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 | 6.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 0.7 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.2 | 1.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 2.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 5.4 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 1.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.2 | 0.9 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 1.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 2.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 1.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 1.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 2.0 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.5 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.2 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 2.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.2 | 1.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 2.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 7.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 2.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.2 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.2 | 2.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 2.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.7 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 1.9 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.7 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.4 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 0.5 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 1.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.6 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 4.5 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.9 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 5.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.6 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 1.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:0021747 | synaptic vesicle targeting(GO:0016080) cochlear nucleus development(GO:0021747) |
| 0.1 | 0.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 2.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.3 | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.1 | 0.6 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.1 | 0.9 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 2.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 1.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 3.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.6 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.1 | 3.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 0.8 | GO:0033292 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 1.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.3 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 1.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.3 | GO:0035936 | testosterone secretion(GO:0035936) negative regulation of steroid hormone secretion(GO:2000832) regulation of testosterone secretion(GO:2000843) |
| 0.1 | 0.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.6 | GO:0042985 | negative regulation of superoxide anion generation(GO:0032929) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.7 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.3 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.2 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.4 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 4.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 0.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 1.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.5 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.4 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.7 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.3 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.9 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.7 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 1.0 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.8 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.5 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.8 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.3 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 1.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 2.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 2.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.7 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.0 | GO:0061314 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 1.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.6 | 7.8 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 1.4 | 5.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.1 | 3.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.1 | 6.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 3.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.7 | 10.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 1.2 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.4 | 6.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 2.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.4 | 1.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.4 | 4.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 1.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 11.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 3.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 3.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 4.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 0.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 3.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 13.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 0.6 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 1.3 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 3.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 3.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 11.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 5.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 12.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 5.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 5.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.4 | 8.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.1 | 6.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.0 | 4.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.9 | 2.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.8 | 10.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.7 | 2.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.7 | 2.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 1.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.5 | 2.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.5 | 2.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 5.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 1.8 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.4 | 8.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 1.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 2.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 1.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.4 | 1.4 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.4 | 1.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 7.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 3.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 2.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.3 | 2.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 1.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 5.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 8.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.8 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 1.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 2.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 7.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 0.5 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.2 | 1.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 6.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 6.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 1.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.6 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 1.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 3.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 5.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.5 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 7.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.3 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 7.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 3.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 10.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 6.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 10.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 5.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 6.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 6.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 4.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 11.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 14.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 2.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 4.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 5.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 3.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 5.9 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 7.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 2.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 2.6 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |