Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
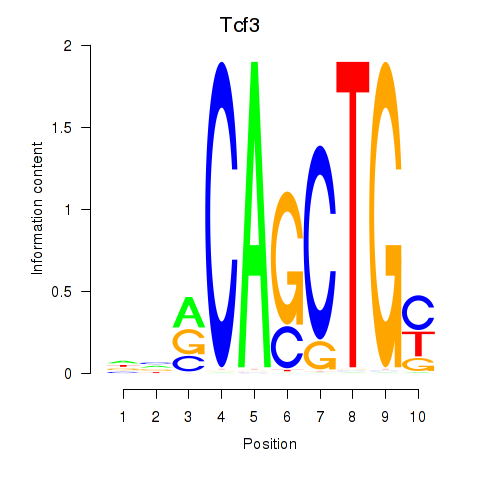
Results for Tcf3
Z-value: 1.59
Transcription factors associated with Tcf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf3
|
ENSMUSG00000020167.15 | transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf3 | mm39_v1_chr10_-_80269436_80269488 | 0.72 | 8.4e-07 | Click! |
Activity profile of Tcf3 motif
Sorted Z-values of Tcf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_112417633 | 19.57 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr2_+_131028861 | 16.81 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr7_-_142223662 | 16.68 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr6_+_30639217 | 16.49 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_-_41291634 | 15.05 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr2_-_28453374 | 13.34 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr11_-_102255999 | 12.72 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr6_+_29694181 | 12.23 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr8_+_23629080 | 11.74 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr8_-_106660470 | 11.64 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr6_-_41354538 | 11.42 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr12_+_109425769 | 11.26 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr17_-_26420300 | 10.90 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr3_-_20329823 | 10.71 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_-_41423004 | 8.94 |
ENSMUST00000095999.7
|
Gm10334
|
predicted gene 10334 |
| chr6_+_41435846 | 8.80 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr6_+_41369290 | 8.58 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr7_+_44866635 | 8.58 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr17_-_26420332 | 8.44 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr8_+_23629046 | 8.42 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr19_-_15901919 | 8.38 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr13_-_55676334 | 7.86 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr1_-_75482975 | 7.38 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr4_-_43523388 | 7.21 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr8_+_95703728 | 7.13 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_21874802 | 7.12 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr4_-_133600308 | 6.97 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_-_43523595 | 6.29 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr3_-_100396635 | 6.29 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr6_+_41331039 | 6.15 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr11_+_115790768 | 6.12 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chrX_-_138772383 | 5.80 |
ENSMUST00000033811.14
ENSMUST00000087401.12 |
Morc4
|
microrchidia 4 |
| chr11_+_53410697 | 5.63 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr5_+_123214332 | 5.63 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr4_+_140737955 | 5.29 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr10_+_128745214 | 5.14 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr8_+_3715747 | 5.11 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1 |
| chr4_-_43523745 | 5.03 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr5_-_113968483 | 5.02 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr18_+_34973605 | 4.98 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr2_+_103800553 | 4.77 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr11_+_104468107 | 4.68 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr2_+_103800459 | 4.54 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr11_-_46203047 | 4.49 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_+_141995545 | 4.47 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_-_16790594 | 4.46 |
ENSMUST00000037762.11
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr2_+_84564394 | 4.42 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr11_+_115790951 | 4.40 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr11_+_104467791 | 4.38 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr1_-_167221344 | 4.24 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr7_+_142025817 | 4.22 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr7_+_142025575 | 4.16 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chrX_+_70599524 | 4.08 |
ENSMUST00000072699.13
ENSMUST00000114582.9 ENSMUST00000015361.11 ENSMUST00000088874.10 |
Hmgb3
|
high mobility group box 3 |
| chr11_+_11636213 | 3.99 |
ENSMUST00000076700.11
ENSMUST00000048122.13 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr9_-_107863062 | 3.98 |
ENSMUST00000048568.6
|
Inka1
|
inka box actin regulator 1 |
| chr4_-_133694607 | 3.93 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr6_-_39397334 | 3.92 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr4_-_133599616 | 3.91 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_-_69496655 | 3.82 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr3_+_95496270 | 3.80 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr14_+_70694887 | 3.79 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr8_+_95720864 | 3.75 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_+_61847589 | 3.67 |
ENSMUST00000201671.4
ENSMUST00000202178.4 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_+_78810919 | 3.67 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr3_+_95496239 | 3.65 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr8_-_123425805 | 3.63 |
ENSMUST00000127984.9
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr7_+_27259895 | 3.56 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_+_121888520 | 3.54 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr1_-_171061838 | 3.49 |
ENSMUST00000193973.2
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr1_+_135060431 | 3.48 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr4_-_133694543 | 3.46 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_95703506 | 3.39 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_95721019 | 3.32 |
ENSMUST00000212976.2
ENSMUST00000212995.2 |
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chrX_-_47551990 | 3.27 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr19_+_5118103 | 3.27 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr7_+_19024994 | 3.26 |
ENSMUST00000108468.5
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr3_+_146110387 | 3.18 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr11_+_32155415 | 3.17 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr8_+_46338498 | 3.12 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr8_-_123187406 | 3.12 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chrX_-_149595711 | 3.12 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chr13_+_97377604 | 3.03 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr11_+_76889415 | 3.02 |
ENSMUST00000108402.9
ENSMUST00000021195.11 |
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr2_-_113883285 | 3.01 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr3_-_151871867 | 2.99 |
ENSMUST00000046614.10
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr2_+_130119077 | 2.98 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr11_+_32155483 | 2.98 |
ENSMUST00000121182.2
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr13_-_56326695 | 2.97 |
ENSMUST00000225063.2
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chrX_-_149595873 | 2.97 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr6_-_68713748 | 2.97 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr7_-_24705320 | 2.96 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr11_-_53371050 | 2.96 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr7_+_92469100 | 2.93 |
ENSMUST00000107180.8
ENSMUST00000107179.2 |
Rab30
|
RAB30, member RAS oncogene family |
| chr14_-_70415117 | 2.90 |
ENSMUST00000022681.11
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr4_+_130643260 | 2.88 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr4_-_116228921 | 2.87 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_-_43813664 | 2.83 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr5_-_151574620 | 2.81 |
ENSMUST00000038131.10
|
Rfc3
|
replication factor C (activator 1) 3 |
| chr8_+_36561982 | 2.81 |
ENSMUST00000110492.2
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr7_-_80037153 | 2.78 |
ENSMUST00000206728.2
|
Fes
|
feline sarcoma oncogene |
| chr12_-_79054050 | 2.75 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chrX_-_36253309 | 2.73 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr7_-_44180700 | 2.71 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr13_-_56326511 | 2.71 |
ENSMUST00000169652.3
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr4_-_131695135 | 2.71 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr11_+_69856222 | 2.70 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr1_+_135060994 | 2.68 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr15_-_78657640 | 2.66 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_-_100036792 | 2.66 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr11_+_61847622 | 2.66 |
ENSMUST00000202389.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr8_+_72050292 | 2.65 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr11_-_107607343 | 2.62 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr5_+_21748523 | 2.60 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr2_-_122441719 | 2.60 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr4_-_93223746 | 2.59 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr1_-_171061902 | 2.57 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr12_+_112978051 | 2.57 |
ENSMUST00000223502.2
ENSMUST00000084891.5 ENSMUST00000220541.2 |
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr7_+_141996067 | 2.53 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr6_+_40941688 | 2.53 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr5_-_140612993 | 2.52 |
ENSMUST00000199157.2
|
Ttyh3
|
tweety family member 3 |
| chr4_+_132968082 | 2.51 |
ENSMUST00000030677.7
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr16_+_87350202 | 2.50 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr16_+_49620883 | 2.45 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr8_+_46338557 | 2.41 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr11_+_69806866 | 2.40 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr15_-_36598263 | 2.39 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_+_26519203 | 2.38 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr17_+_56610321 | 2.34 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chrX_-_56384089 | 2.32 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr9_+_21249118 | 2.30 |
ENSMUST00000034697.8
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr1_+_135764092 | 2.24 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr10_+_126899468 | 2.21 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr2_+_156681991 | 2.20 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr19_+_56536685 | 2.18 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr9_-_39515420 | 2.18 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr1_+_75336965 | 2.13 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr2_-_125348305 | 2.08 |
ENSMUST00000028633.13
|
Fbn1
|
fibrillin 1 |
| chr7_+_44866095 | 2.07 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr7_-_105131407 | 2.06 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr17_+_56610396 | 2.03 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr4_-_131664478 | 2.02 |
ENSMUST00000155990.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr19_+_6326755 | 1.98 |
ENSMUST00000025684.4
|
Ehd1
|
EH-domain containing 1 |
| chr5_-_73413888 | 1.96 |
ENSMUST00000101127.12
|
Fryl
|
FRY like transcription coactivator |
| chr7_+_28488380 | 1.95 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr11_+_63019799 | 1.94 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr7_+_126811831 | 1.93 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr19_+_53128901 | 1.93 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr4_-_137137088 | 1.92 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr9_+_107468146 | 1.90 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr2_+_25152600 | 1.89 |
ENSMUST00000114336.4
|
Tprn
|
taperin |
| chr13_-_111945499 | 1.88 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr10_-_128755127 | 1.86 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr2_-_167503416 | 1.86 |
ENSMUST00000125544.3
ENSMUST00000006587.7 |
Gm20431
Tmem189
|
predicted gene 20431 transmembrane protein 189 |
| chr5_+_35146727 | 1.85 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr9_-_40257586 | 1.84 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr5_-_137530214 | 1.81 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr6_-_68609426 | 1.80 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr2_-_91854844 | 1.79 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr11_+_96820091 | 1.78 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr11_+_3152874 | 1.75 |
ENSMUST00000179770.8
ENSMUST00000110048.8 |
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr15_-_82128538 | 1.73 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr7_-_80037622 | 1.73 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr7_-_126224848 | 1.72 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr12_-_114672701 | 1.71 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr17_+_75485791 | 1.70 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_35413415 | 1.70 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr8_+_106245368 | 1.69 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr15_-_82128888 | 1.67 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr19_-_6065181 | 1.67 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chr6_-_85351524 | 1.67 |
ENSMUST00000060837.10
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I) |
| chr2_+_124910037 | 1.66 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr19_-_56536646 | 1.66 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr2_+_118943274 | 1.65 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr7_+_101060093 | 1.64 |
ENSMUST00000084894.15
|
Gm45837
|
predicted gene 45837 |
| chr11_+_99748741 | 1.63 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr2_-_164621641 | 1.62 |
ENSMUST00000103095.5
|
Tnnc2
|
troponin C2, fast |
| chr7_+_19144950 | 1.62 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr1_-_182929025 | 1.61 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr6_-_69792108 | 1.61 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr7_+_24982206 | 1.61 |
ENSMUST00000165239.3
|
Cic
|
capicua transcriptional repressor |
| chr5_+_35146880 | 1.59 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_+_115705550 | 1.58 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr13_+_37529184 | 1.56 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr7_+_3339077 | 1.56 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_+_70548622 | 1.56 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr19_+_32463151 | 1.55 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr5_-_114911548 | 1.55 |
ENSMUST00000178440.8
ENSMUST00000043283.14 ENSMUST00000112185.9 ENSMUST00000155908.8 |
Git2
|
GIT ArfGAP 2 |
| chr7_+_3339059 | 1.55 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr8_-_106198112 | 1.54 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_-_141579689 | 1.54 |
ENSMUST00000209732.2
|
Mob2
|
MOB kinase activator 2 |
| chr14_-_30329765 | 1.53 |
ENSMUST00000112207.8
ENSMUST00000112206.8 ENSMUST00000112202.8 ENSMUST00000112203.2 |
Prkcd
|
protein kinase C, delta |
| chrX_+_70600481 | 1.53 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr3_-_75177378 | 1.52 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr16_+_57173456 | 1.51 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr16_+_35590745 | 1.51 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr11_-_120538928 | 1.50 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_+_67602565 | 1.49 |
ENSMUST00000005671.10
|
Igf1r
|
insulin-like growth factor I receptor |
| chr2_-_92222979 | 1.49 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chrX_+_168662592 | 1.47 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr6_-_68812291 | 1.46 |
ENSMUST00000199143.5
ENSMUST00000103335.3 |
Igkv12-89
|
immunoglobulin kappa chain variable 12-89 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 16.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.3 | 4.0 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.3 | 5.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 1.2 | 3.7 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.2 | 10.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.1 | 5.7 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 1.0 | 8.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.0 | 5.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.9 | 5.6 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.9 | 5.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 5.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.8 | 2.4 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.8 | 3.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.8 | 6.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.8 | 3.0 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.7 | 11.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.7 | 0.7 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) |
| 0.7 | 17.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.7 | 3.4 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.6 | 1.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.6 | 7.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.6 | 4.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.6 | 2.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 1.7 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.6 | 2.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.6 | 1.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.6 | 1.7 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.5 | 9.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 | 1.6 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.5 | 2.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 | 2.1 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.5 | 3.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 2.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.5 | 1.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.5 | 2.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.5 | 1.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.5 | 1.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 7.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.5 | 13.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 7.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.5 | 1.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.5 | 1.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.4 | 4.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 1.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 1.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.4 | 10.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.4 | 1.7 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.4 | 18.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.4 | 64.2 | GO:0007586 | digestion(GO:0007586) |
| 0.4 | 1.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.4 | 1.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 3.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.4 | 3.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 1.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.4 | 1.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.4 | 3.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.4 | 1.5 | GO:1903944 | estrous cycle(GO:0044849) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.4 | 3.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.4 | 3.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.4 | 1.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 2.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 1.4 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.3 | 1.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 2.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.3 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 1.0 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.3 | 2.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 0.6 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 11.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.3 | 2.8 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 0.9 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 | 2.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 1.8 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 1.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.3 | 0.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 4.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 1.2 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.3 | 1.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.3 | 1.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 2.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 0.6 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.3 | 7.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.3 | 4.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.3 | 3.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 4.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 2.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.0 | GO:0090096 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 2.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 1.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 9.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 1.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 3.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.2 | 0.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.2 | 1.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 1.3 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.2 | 1.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 1.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 0.8 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 1.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 6.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 0.8 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.4 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 0.6 | GO:0099542 | retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 2.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 1.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 5.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 2.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 2.8 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.1 | 0.9 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 2.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.3 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.1 | 0.3 | GO:2000554 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.1 | 0.3 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 2.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 4.5 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 1.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 2.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.3 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) |
| 0.1 | 1.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 0.9 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 | 0.5 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 2.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 2.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.4 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.1 | 2.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.5 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.5 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 2.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 2.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 2.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.3 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 2.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 2.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 3.3 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 6.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 1.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.2 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.5 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.5 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 10.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 2.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 1.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 2.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 5.7 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 0.5 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 2.4 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 3.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 5.1 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.0 | 6.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 1.7 | 5.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.6 | 17.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.2 | 18.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.0 | 12.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.0 | 3.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.9 | 2.8 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.8 | 3.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.7 | 6.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 2.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.7 | 12.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 17.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 7.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 1.5 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.5 | 1.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 2.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 2.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.4 | 6.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 13.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 3.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 2.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 7.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 2.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.9 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.2 | 1.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 4.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 6.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 2.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 10.0 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 2.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 3.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 11.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 12.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 18.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 4.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 2.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 6.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 3.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 9.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 2.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 6.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 6.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 4.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 15.2 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 53.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 6.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 2.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 3.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 2.1 | 19.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.8 | 18.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.2 | 6.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 1.1 | 5.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.0 | 3.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.9 | 3.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.9 | 27.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.9 | 2.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 8.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.8 | 2.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.6 | 4.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.6 | 6.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 1.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 12.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 2.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.5 | 32.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 10.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.4 | 1.3 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.4 | 1.6 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.3 | 1.4 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.3 | 1.4 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 1.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 3.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.3 | 1.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 2.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 76.1 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.3 | 2.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 1.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 20.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 1.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 2.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 1.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 2.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 3.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 2.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 7.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 5.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 10.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 1.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 7.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 6.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 2.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 2.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 6.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 1.7 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 4.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 8.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 14.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 11.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 3.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 5.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 4.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 3.3 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 16.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 9.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 13.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 9.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 11.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 4.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 16.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 6.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 9.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 7.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 19.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 5.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 5.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.8 | 24.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 39.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 6.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 17.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 8.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 11.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 4.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 10.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 9.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 4.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 3.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 1.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 5.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 5.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 13.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.6 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |