Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
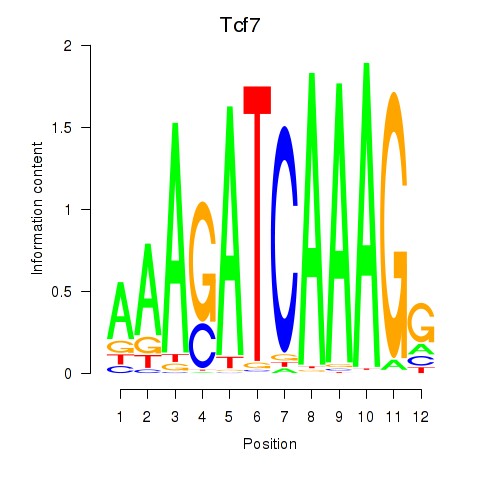
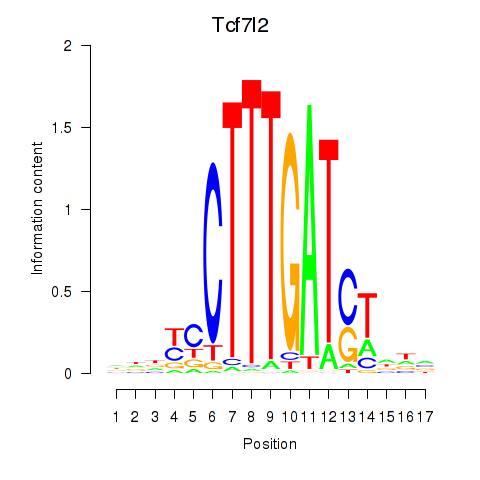
Results for Tcf7_Tcf7l2
Z-value: 0.72
Transcription factors associated with Tcf7_Tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7
|
ENSMUSG00000000782.17 | transcription factor 7, T cell specific |
|
Tcf7l2
|
ENSMUSG00000024985.21 | transcription factor 7 like 2, T cell specific, HMG box |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7 | mm39_v1_chr11_-_52174129_52174221 | 0.56 | 3.8e-04 | Click! |
| Tcf7l2 | mm39_v1_chr19_+_55886708_55886741 | 0.49 | 2.7e-03 | Click! |
Activity profile of Tcf7_Tcf7l2 motif
Sorted Z-values of Tcf7_Tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_145816774 | 4.74 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr2_+_102536701 | 3.93 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_88093726 | 3.55 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr10_-_127206300 | 3.48 |
ENSMUST00000026472.10
|
Inhbc
|
inhibin beta-C |
| chr7_+_13467422 | 3.40 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr10_-_95678748 | 3.01 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr3_+_118355778 | 2.95 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr10_-_95678786 | 2.93 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr1_-_139487951 | 2.91 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr7_+_13357892 | 2.43 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr6_+_115398996 | 2.39 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr1_-_139708906 | 2.33 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr3_+_118355811 | 2.24 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr19_+_39980868 | 2.18 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr7_+_25760922 | 2.16 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr2_-_154916367 | 2.14 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr4_+_45848918 | 1.96 |
ENSMUST00000030011.6
|
Stra6l
|
STRA6-like |
| chr6_-_47790272 | 1.94 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr9_+_46151994 | 1.88 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr13_+_24511387 | 1.85 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr8_-_25066313 | 1.79 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr5_-_145406533 | 1.77 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr16_+_22739028 | 1.73 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr8_-_112356957 | 1.63 |
ENSMUST00000070004.4
|
Ldhd
|
lactate dehydrogenase D |
| chr5_-_87716882 | 1.61 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr19_+_58658779 | 1.61 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr10_-_115423644 | 1.60 |
ENSMUST00000020350.15
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr8_-_112417633 | 1.58 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chrX_+_59044796 | 1.57 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr7_+_67305162 | 1.55 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr14_-_34077340 | 1.50 |
ENSMUST00000052126.6
|
Fam25c
|
family with sequence similarity 25, member C |
| chr11_-_52174129 | 1.42 |
ENSMUST00000109071.3
|
Tcf7
|
transcription factor 7, T cell specific |
| chr15_-_54141816 | 1.42 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr13_-_56696310 | 1.40 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr9_+_77829191 | 1.39 |
ENSMUST00000133757.8
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr13_-_56696222 | 1.38 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr19_+_58658838 | 1.37 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr19_+_44980565 | 1.37 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr6_+_117145496 | 1.35 |
ENSMUST00000112866.8
ENSMUST00000112871.8 ENSMUST00000073043.5 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr7_-_30776081 | 1.33 |
ENSMUST00000072331.13
ENSMUST00000167369.8 |
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr2_-_34990689 | 1.30 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr1_+_87998487 | 1.25 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr19_+_4771089 | 1.21 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr3_-_113166153 | 1.15 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr2_+_22958179 | 1.14 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr12_-_84455764 | 1.12 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr1_+_128171859 | 1.10 |
ENSMUST00000027592.6
|
Ubxn4
|
UBX domain protein 4 |
| chr1_+_157334298 | 1.09 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334347 | 1.08 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr10_+_75242745 | 1.07 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr15_+_10981833 | 1.06 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr17_+_79922329 | 1.05 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr13_-_41981812 | 1.05 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr7_+_132460954 | 1.05 |
ENSMUST00000084497.12
ENSMUST00000106161.8 |
Abraxas2
|
BRISC complex subunit |
| chr17_-_35077089 | 1.03 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr13_-_41981893 | 1.03 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_79922486 | 1.03 |
ENSMUST00000225357.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr18_+_84106188 | 1.01 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr10_-_24803336 | 1.00 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr14_-_70561231 | 0.99 |
ENSMUST00000151011.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr6_+_54244116 | 0.97 |
ENSMUST00000114402.9
|
Chn2
|
chimerin 2 |
| chr10_+_89580849 | 0.96 |
ENSMUST00000020112.7
|
Uhrf1bp1l
|
UHRF1 (ICBP90) binding protein 1-like |
| chr9_-_15212745 | 0.96 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr12_+_87193922 | 0.95 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr13_+_24054267 | 0.95 |
ENSMUST00000006785.8
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr7_-_144493560 | 0.93 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr2_-_34951443 | 0.92 |
ENSMUST00000028233.7
|
Hc
|
hemolytic complement |
| chr10_+_61484331 | 0.91 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr9_-_15212849 | 0.89 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr6_+_138117295 | 0.88 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr6_+_138117519 | 0.87 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr6_+_54016543 | 0.87 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr14_-_45456821 | 0.86 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr3_+_57332735 | 0.85 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr1_+_87983189 | 0.85 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_+_122277372 | 0.84 |
ENSMUST00000197073.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr11_-_59927688 | 0.84 |
ENSMUST00000102692.10
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr2_+_14234198 | 0.82 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr15_+_31224460 | 0.81 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr8_+_36924702 | 0.81 |
ENSMUST00000135373.8
ENSMUST00000147525.9 |
Trmt9b
|
tRNA methyltransferase 9B |
| chr7_-_34914675 | 0.81 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr4_-_19922599 | 0.79 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr3_-_53771185 | 0.79 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr1_+_87983099 | 0.79 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr13_+_47276132 | 0.78 |
ENSMUST00000068891.12
|
Rnf144b
|
ring finger protein 144B |
| chr9_+_106325828 | 0.78 |
ENSMUST00000217496.2
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr11_+_108811626 | 0.78 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr9_-_122694999 | 0.75 |
ENSMUST00000214558.2
ENSMUST00000216721.2 |
Zfp445
|
zinc finger protein 445 |
| chr7_-_101494472 | 0.74 |
ENSMUST00000211566.2
ENSMUST00000094141.7 ENSMUST00000209329.2 |
Folr2
|
folate receptor 2 (fetal) |
| chr9_-_96634874 | 0.72 |
ENSMUST00000152594.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_+_70305267 | 0.72 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr9_-_96601574 | 0.71 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_+_22958956 | 0.71 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr4_-_63072367 | 0.69 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr5_+_138083345 | 0.69 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr6_+_21985902 | 0.69 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr9_+_106325860 | 0.69 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr8_+_96078886 | 0.67 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr2_+_20742115 | 0.67 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr14_-_61283911 | 0.65 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_-_45626237 | 0.65 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr11_+_98938137 | 0.64 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr5_+_91039092 | 0.64 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr4_+_148686985 | 0.62 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr5_-_105387395 | 0.61 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr3_+_122305819 | 0.61 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr11_-_99213769 | 0.61 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr4_+_134124691 | 0.60 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr11_+_108271990 | 0.59 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr1_-_59276252 | 0.58 |
ENSMUST00000163058.2
ENSMUST00000160945.2 ENSMUST00000027178.13 |
Als2
|
alsin Rho guanine nucleotide exchange factor |
| chr6_+_78382131 | 0.58 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr1_+_182782335 | 0.57 |
ENSMUST00000193687.7
|
Tlr5
|
toll-like receptor 5 |
| chr6_-_148732893 | 0.56 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr10_+_127685785 | 0.55 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr7_+_37883300 | 0.55 |
ENSMUST00000179992.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr7_+_37883216 | 0.55 |
ENSMUST00000177983.2
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr11_+_114743044 | 0.55 |
ENSMUST00000142262.2
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr14_+_26959975 | 0.54 |
ENSMUST00000049206.6
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr9_-_50657800 | 0.54 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr10_-_94780695 | 0.54 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr10_-_128425519 | 0.54 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr2_+_91033230 | 0.54 |
ENSMUST00000150403.8
ENSMUST00000002172.14 ENSMUST00000238832.2 ENSMUST00000239169.2 ENSMUST00000155418.2 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr4_-_124744266 | 0.54 |
ENSMUST00000137769.3
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr9_-_86577940 | 0.54 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr11_+_102036356 | 0.53 |
ENSMUST00000055409.6
|
Nags
|
N-acetylglutamate synthase |
| chr9_-_122695071 | 0.53 |
ENSMUST00000216063.2
|
Zfp445
|
zinc finger protein 445 |
| chr7_-_79492091 | 0.52 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr19_+_8816663 | 0.52 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr5_+_65288418 | 0.52 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr13_+_33187205 | 0.52 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr4_+_141095415 | 0.52 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr4_+_40723083 | 0.52 |
ENSMUST00000149794.2
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr6_-_148732946 | 0.52 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr13_-_104056803 | 0.51 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr3_+_137377285 | 0.51 |
ENSMUST00000166899.3
|
Gm21962
|
predicted gene, 21962 |
| chr18_+_61688378 | 0.50 |
ENSMUST00000165721.8
ENSMUST00000115246.9 ENSMUST00000166990.8 ENSMUST00000163205.8 ENSMUST00000170862.8 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr5_+_143220897 | 0.50 |
ENSMUST00000077485.11
ENSMUST00000032591.15 |
Zfp12
|
zinc finger protein 12 |
| chr2_+_30156733 | 0.50 |
ENSMUST00000113645.8
ENSMUST00000133877.8 ENSMUST00000139719.8 ENSMUST00000113643.8 ENSMUST00000150695.8 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr14_+_56255422 | 0.50 |
ENSMUST00000022836.6
|
Mcpt1
|
mast cell protease 1 |
| chr1_+_88154727 | 0.50 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr19_+_38121248 | 0.49 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr10_-_128204545 | 0.49 |
ENSMUST00000220027.2
|
Coq10a
|
coenzyme Q10A |
| chr5_+_118114795 | 0.49 |
ENSMUST00000202447.4
|
Fbxo21
|
F-box protein 21 |
| chr9_-_65734826 | 0.49 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609 |
| chr3_+_85946145 | 0.48 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr12_-_79219382 | 0.48 |
ENSMUST00000055262.13
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr5_-_97259302 | 0.48 |
ENSMUST00000196078.5
|
Paqr3
|
progestin and adipoQ receptor family member III |
| chr13_+_24560052 | 0.48 |
ENSMUST00000110391.4
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr17_-_74257164 | 0.48 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr10_-_128204806 | 0.48 |
ENSMUST00000043211.7
ENSMUST00000220227.2 |
Coq10a
|
coenzyme Q10A |
| chr13_+_54738014 | 0.47 |
ENSMUST00000026986.7
|
Higd2a
|
HIG1 domain family, member 2A |
| chr1_+_157286124 | 0.47 |
ENSMUST00000193791.6
ENSMUST00000046743.11 ENSMUST00000119891.7 |
Cryzl2
|
crystallin zeta like 2 |
| chr5_+_118114826 | 0.47 |
ENSMUST00000035579.10
|
Fbxo21
|
F-box protein 21 |
| chr5_-_147013384 | 0.47 |
ENSMUST00000016664.8
|
Lnx2
|
ligand of numb-protein X 2 |
| chr14_+_12016304 | 0.47 |
ENSMUST00000161302.8
|
Fhit
|
fragile histidine triad gene |
| chr6_+_34722926 | 0.47 |
ENSMUST00000126181.8
|
Cald1
|
caldesmon 1 |
| chr10_-_22696025 | 0.46 |
ENSMUST00000218002.2
ENSMUST00000049930.9 |
Tcf21
|
transcription factor 21 |
| chr14_+_54669054 | 0.45 |
ENSMUST00000089688.6
ENSMUST00000225641.2 |
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr19_+_38121214 | 0.45 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr6_-_124718316 | 0.45 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr2_+_134627987 | 0.45 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr15_+_9436114 | 0.45 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chr18_-_66155651 | 0.45 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr2_+_71884943 | 0.44 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_-_43959993 | 0.44 |
ENSMUST00000137557.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr6_+_21986445 | 0.44 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr18_-_12996348 | 0.44 |
ENSMUST00000143077.8
ENSMUST00000117361.8 ENSMUST00000234871.2 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr2_+_30156523 | 0.44 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr10_+_71183571 | 0.43 |
ENSMUST00000079252.13
|
Ipmk
|
inositol polyphosphate multikinase |
| chr10_+_38841511 | 0.43 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr7_+_24310738 | 0.43 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr17_+_44263890 | 0.42 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr9_-_64248426 | 0.42 |
ENSMUST00000120760.8
ENSMUST00000168844.9 |
Dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr4_+_99082373 | 0.41 |
ENSMUST00000180278.2
|
Atg4c
|
autophagy related 4C, cysteine peptidase |
| chr7_-_13723513 | 0.41 |
ENSMUST00000165167.8
ENSMUST00000108520.4 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr11_+_99755302 | 0.41 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr6_+_34722887 | 0.40 |
ENSMUST00000123823.8
ENSMUST00000136907.8 |
Cald1
|
caldesmon 1 |
| chr10_-_8761777 | 0.39 |
ENSMUST00000015449.6
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr14_+_102078038 | 0.39 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr4_+_95855442 | 0.39 |
ENSMUST00000030306.14
|
Hook1
|
hook microtubule tethering protein 1 |
| chr10_-_122912272 | 0.39 |
ENSMUST00000219203.2
ENSMUST00000073792.11 |
Mon2
|
MON2 homolog, regulator of endosome to Golgi trafficking |
| chr10_+_21854540 | 0.38 |
ENSMUST00000142174.8
ENSMUST00000164659.8 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_59038031 | 0.38 |
ENSMUST00000091112.6
ENSMUST00000065220.13 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr10_+_96453408 | 0.38 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr18_+_77031788 | 0.38 |
ENSMUST00000097522.11
ENSMUST00000097521.12 ENSMUST00000145634.9 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr1_-_171910324 | 0.37 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr8_+_45960931 | 0.37 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_-_96721829 | 0.37 |
ENSMUST00000047702.8
|
Cd160
|
CD160 antigen |
| chr14_+_63673843 | 0.37 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr6_-_85684840 | 0.37 |
ENSMUST00000179613.2
|
Nat8f7
|
N-acetyltransferase 8 (GCN5-related) family member 7 |
| chr10_-_60983438 | 0.36 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr2_-_38816229 | 0.36 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr2_+_163348728 | 0.36 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr14_+_52122439 | 0.35 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr10_+_29189159 | 0.34 |
ENSMUST00000160399.8
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr5_-_44259374 | 0.34 |
ENSMUST00000171543.8
|
Prom1
|
prominin 1 |
| chr19_+_56276343 | 0.33 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr5_-_44259293 | 0.33 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr13_-_108026590 | 0.33 |
ENSMUST00000225822.4
ENSMUST00000225197.3 |
Zswim6
|
zinc finger SWIM-type containing 6 |
| chr10_-_30494431 | 0.33 |
ENSMUST00000161074.8
|
Hint3
|
histidine triad nucleotide binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7_Tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.7 | 3.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.7 | 2.0 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.6 | 1.9 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.6 | 2.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.6 | 2.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.6 | 3.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 6.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.4 | 1.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 2.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 1.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 2.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.6 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 1.2 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 1.0 | GO:0090467 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 3.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.8 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.2 | 0.8 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.5 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.4 | GO:0060466 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to fluoride(GO:1902617) |
| 0.1 | 1.4 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 1.4 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 4.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.1 | 0.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.4 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.1 | 1.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 1.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 1.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 1.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.2 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.2 | GO:0042197 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:2000331 | positive regulation of Wnt protein secretion(GO:0061357) regulation of terminal button organization(GO:2000331) |
| 0.1 | 1.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.3 | GO:0060983 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 0.1 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.5 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.5 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.2 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 3.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 1.1 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.7 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 1.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 1.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.1 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.1 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 1.6 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 1.4 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 0.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 2.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 2.6 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.5 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0098967 | synaptic vesicle docking(GO:0016081) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 2.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 2.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 3.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 6.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 6.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.8 | 2.3 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.8 | 6.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.7 | 3.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 1.8 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.5 | 1.6 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.5 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 2.8 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 1.0 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.3 | 2.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 1.0 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 1.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 1.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 0.5 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 2.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.5 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 1.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 3.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 6.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 1.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 0.5 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 4.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.9 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.5 | GO:0043546 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.2 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.1 | 0.2 | GO:0016824 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.2 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.1 | 3.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 1.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 1.0 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.7 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 6.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.2 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 3.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.0 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |