Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2d
Z-value: 1.19
Transcription factors associated with Tfap2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2d
|
ENSMUSG00000042596.8 | transcription factor AP-2, delta |
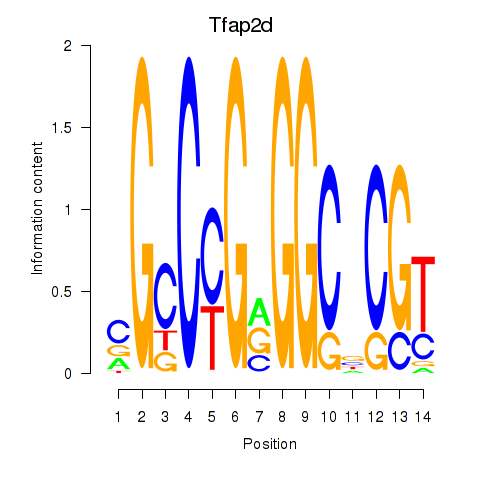
Activity profile of Tfap2d motif
Sorted Z-values of Tfap2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_57143437 | 6.67 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr10_-_88339935 | 6.59 |
ENSMUST00000117440.8
|
Chpt1
|
choline phosphotransferase 1 |
| chrX_+_93278588 | 5.68 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chrX_+_93278526 | 5.36 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr15_-_31367668 | 5.06 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr10_-_88339773 | 4.95 |
ENSMUST00000117579.8
ENSMUST00000073783.6 |
Chpt1
|
choline phosphotransferase 1 |
| chr2_-_173117936 | 4.92 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr10_-_88339814 | 4.75 |
ENSMUST00000020253.15
|
Chpt1
|
choline phosphotransferase 1 |
| chr2_+_167530835 | 4.28 |
ENSMUST00000070642.4
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr2_+_27567213 | 4.27 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chrX_+_93278203 | 4.13 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr8_+_86219191 | 4.08 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr7_+_65511777 | 3.91 |
ENSMUST00000055576.12
ENSMUST00000098391.11 |
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr5_-_34345014 | 3.86 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr7_-_34914675 | 3.59 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr17_+_37253802 | 3.50 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr4_-_46991842 | 3.45 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr10_+_61531282 | 3.29 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr12_-_30423356 | 3.27 |
ENSMUST00000021004.14
|
Sntg2
|
syntrophin, gamma 2 |
| chr4_-_41640321 | 3.20 |
ENSMUST00000127306.2
|
Enho
|
energy homeostasis associated |
| chr15_-_31367872 | 3.07 |
ENSMUST00000123325.9
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr17_-_24428351 | 2.95 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr2_+_155907658 | 2.84 |
ENSMUST00000137966.2
|
Spag4
|
sperm associated antigen 4 |
| chr7_+_65511482 | 2.78 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr1_-_39616445 | 2.75 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr2_-_26096547 | 2.73 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr8_-_121361097 | 2.69 |
ENSMUST00000123927.9
|
1190005I06Rik
|
RIKEN cDNA 1190005I06 gene |
| chr4_-_108637700 | 2.68 |
ENSMUST00000106658.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr13_+_119565424 | 2.60 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr6_-_124391994 | 2.57 |
ENSMUST00000035861.6
ENSMUST00000112532.8 ENSMUST00000080557.12 |
Pex5
|
peroxisomal biogenesis factor 5 |
| chr11_-_72686853 | 2.52 |
ENSMUST00000156294.8
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr2_-_173118315 | 2.51 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_+_141095415 | 2.49 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr6_+_72332423 | 2.49 |
ENSMUST00000069695.9
ENSMUST00000132243.3 |
Tmem150a
|
transmembrane protein 150A |
| chr6_-_48422447 | 2.40 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr4_+_40722911 | 2.32 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr8_+_36924702 | 2.31 |
ENSMUST00000135373.8
ENSMUST00000147525.9 |
Trmt9b
|
tRNA methyltransferase 9B |
| chr6_+_72332449 | 2.25 |
ENSMUST00000206064.2
|
Tmem150a
|
transmembrane protein 150A |
| chr2_+_27567246 | 2.24 |
ENSMUST00000166775.8
|
Rxra
|
retinoid X receptor alpha |
| chr5_+_34445944 | 2.20 |
ENSMUST00000207754.2
|
Cfap99
|
cilia and flagella associated protein 99 |
| chr14_+_73790105 | 2.20 |
ENSMUST00000160507.8
ENSMUST00000022706.7 |
Sucla2
|
succinate-Coenzyme A ligase, ADP-forming, beta subunit |
| chr4_+_84802650 | 2.16 |
ENSMUST00000169371.9
|
Cntln
|
centlein, centrosomal protein |
| chr16_-_4238280 | 2.12 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr14_-_20844074 | 2.11 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr15_+_87428483 | 2.09 |
ENSMUST00000230414.2
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr5_+_119808722 | 2.09 |
ENSMUST00000079719.11
|
Tbx3
|
T-box 3 |
| chr5_+_108842294 | 2.06 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr11_-_72686627 | 2.05 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr5_+_114706077 | 2.02 |
ENSMUST00000043650.8
|
Fam222a
|
family with sequence similarity 222, member A |
| chr4_-_108637979 | 2.02 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr5_+_119808890 | 2.02 |
ENSMUST00000121021.8
|
Tbx3
|
T-box 3 |
| chr1_-_39616369 | 2.01 |
ENSMUST00000195705.2
|
Rnf149
|
ring finger protein 149 |
| chr14_-_18817743 | 2.01 |
ENSMUST00000167430.8
|
Gm3020
|
predicted gene 3020 |
| chr5_-_34445662 | 2.00 |
ENSMUST00000094868.10
|
Zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr4_-_115875055 | 1.97 |
ENSMUST00000049095.6
|
Faah
|
fatty acid amide hydrolase |
| chr3_+_40904253 | 1.95 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr2_-_32872032 | 1.94 |
ENSMUST00000195863.2
ENSMUST00000028129.13 |
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8 |
| chr4_+_84802592 | 1.90 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr2_+_167922924 | 1.88 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr17_-_66826661 | 1.88 |
ENSMUST00000167962.2
ENSMUST00000070538.12 |
Rab12
|
RAB12, member RAS oncogene family |
| chr14_-_19420488 | 1.86 |
ENSMUST00000166494.2
|
Gm2897
|
predicted gene 2897 |
| chr8_+_84699580 | 1.84 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr2_-_168072493 | 1.82 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr14_+_14901127 | 1.81 |
ENSMUST00000163790.2
|
Gm3558
|
predicted gene 3558 |
| chr12_+_103498542 | 1.79 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr14_+_16508028 | 1.79 |
ENSMUST00000163885.2
|
Gm3248
|
predicted gene 3248 |
| chr2_-_35091076 | 1.77 |
ENSMUST00000028238.15
|
Rab14
|
RAB14, member RAS oncogene family |
| chr11_+_97690391 | 1.73 |
ENSMUST00000043843.12
|
Lasp1
|
LIM and SH3 protein 1 |
| chr2_+_90677499 | 1.72 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr11_+_108811626 | 1.72 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr14_+_16589391 | 1.71 |
ENSMUST00000164484.8
|
Gm8237
|
predicted gene 8237 |
| chr2_-_32872019 | 1.68 |
ENSMUST00000153484.7
|
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8 |
| chr7_-_79882501 | 1.68 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr5_+_34817704 | 1.66 |
ENSMUST00000074651.11
ENSMUST00000001112.14 |
Grk4
|
G protein-coupled receptor kinase 4 |
| chr6_+_91661034 | 1.65 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr8_-_70355208 | 1.64 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr4_+_84802513 | 1.63 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr14_-_19635203 | 1.63 |
ENSMUST00000170694.9
|
Gm2237
|
predicted gene 2237 |
| chr14_-_31552608 | 1.63 |
ENSMUST00000014640.9
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr2_+_76505619 | 1.62 |
ENSMUST00000111920.2
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr5_+_88712840 | 1.59 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr14_-_19261196 | 1.59 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr16_-_18405709 | 1.59 |
ENSMUST00000232335.2
|
Tbx1
|
T-box 1 |
| chr11_-_107685383 | 1.59 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr7_-_127307898 | 1.57 |
ENSMUST00000207019.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr7_+_106969919 | 1.57 |
ENSMUST00000137663.2
|
Syt9
|
synaptotagmin IX |
| chr9_-_29323500 | 1.56 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr7_+_106969950 | 1.56 |
ENSMUST00000073459.12
|
Syt9
|
synaptotagmin IX |
| chr19_-_7183596 | 1.55 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr2_+_92430043 | 1.55 |
ENSMUST00000065797.7
|
Chst1
|
carbohydrate sulfotransferase 1 |
| chr19_-_37340010 | 1.54 |
ENSMUST00000131070.3
|
Ide
|
insulin degrading enzyme |
| chr10_-_31485180 | 1.53 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr13_+_9326513 | 1.53 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr11_+_97690585 | 1.53 |
ENSMUST00000129558.8
|
Lasp1
|
LIM and SH3 protein 1 |
| chr2_+_179666744 | 1.51 |
ENSMUST00000055485.12
|
Lsm14b
|
LSM family member 14B |
| chr9_-_29323032 | 1.51 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr19_-_10582672 | 1.49 |
ENSMUST00000236478.2
ENSMUST00000236950.2 |
Tkfc
|
triokinase, FMN cyclase |
| chr2_-_35090961 | 1.46 |
ENSMUST00000230751.2
|
Rab14
|
RAB14, member RAS oncogene family |
| chr5_+_28370687 | 1.46 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr2_+_30061469 | 1.46 |
ENSMUST00000015481.6
|
Endog
|
endonuclease G |
| chr19_-_4748696 | 1.45 |
ENSMUST00000225896.2
ENSMUST00000177696.8 |
Gm960
|
predicted gene 960 |
| chr16_-_35589726 | 1.44 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr19_-_7183626 | 1.43 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr17_-_13271183 | 1.43 |
ENSMUST00000091648.4
|
Gpr31b
|
G protein-coupled receptor 31, D17Leh66b region |
| chr14_-_31552335 | 1.43 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr7_-_141055044 | 1.43 |
ENSMUST00000043870.9
|
Polr2l
|
polymerase (RNA) II (DNA directed) polypeptide L |
| chr2_+_155907100 | 1.42 |
ENSMUST00000038860.12
|
Spag4
|
sperm associated antigen 4 |
| chr11_+_72498029 | 1.41 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr19_-_10582773 | 1.41 |
ENSMUST00000237788.2
|
Tkfc
|
triokinase, FMN cyclase |
| chr8_-_105122397 | 1.39 |
ENSMUST00000179802.2
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chrX_-_7440480 | 1.37 |
ENSMUST00000115742.9
ENSMUST00000150787.8 |
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr5_+_141227245 | 1.35 |
ENSMUST00000085774.11
|
Sdk1
|
sidekick cell adhesion molecule 1 |
| chr2_-_35994819 | 1.34 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr15_+_99489018 | 1.32 |
ENSMUST00000229728.2
ENSMUST00000231163.2 |
Aqp5
|
aquaporin 5 |
| chr14_-_19057159 | 1.31 |
ENSMUST00000170123.2
|
Gm10409
|
predicted gene 10409 |
| chr11_+_11065782 | 1.31 |
ENSMUST00000056344.5
|
Vwc2
|
von Willebrand factor C domain containing 2 |
| chr2_-_32178034 | 1.29 |
ENSMUST00000183946.8
ENSMUST00000113400.3 ENSMUST00000050410.11 |
Swi5
|
SWI5 recombination repair homolog (yeast) |
| chr14_+_15295240 | 1.28 |
ENSMUST00000172431.8
|
Gm3512
|
predicted gene 3512 |
| chr5_-_34445751 | 1.28 |
ENSMUST00000114368.2
|
Zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr1_+_74430575 | 1.28 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr3_+_9315662 | 1.27 |
ENSMUST00000155203.2
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr7_-_127308059 | 1.27 |
ENSMUST00000061468.9
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr11_+_69826603 | 1.26 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr4_+_152093260 | 1.26 |
ENSMUST00000097773.4
|
Klhl21
|
kelch-like 21 |
| chr2_+_91087668 | 1.25 |
ENSMUST00000111349.9
ENSMUST00000131711.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_+_37253916 | 1.25 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr10_+_11219117 | 1.24 |
ENSMUST00000069106.5
|
Epm2a
|
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha |
| chr15_+_34837501 | 1.23 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr13_+_31809774 | 1.22 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr4_-_114766070 | 1.20 |
ENSMUST00000068654.5
|
Foxd2
|
forkhead box D2 |
| chr14_+_15579811 | 1.20 |
ENSMUST00000171906.2
|
Gm3667
|
predicted gene 3667 |
| chr16_-_38533597 | 1.20 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr4_+_48279794 | 1.20 |
ENSMUST00000030029.10
|
Invs
|
inversin |
| chr11_+_49916136 | 1.19 |
ENSMUST00000054684.14
ENSMUST00000238748.2 ENSMUST00000102776.5 |
Rnf130
|
ring finger protein 130 |
| chr2_+_122461079 | 1.19 |
ENSMUST00000239506.1
|
SPATA5L1
|
spermatosis associated 5-like 1 |
| chr15_-_82872073 | 1.18 |
ENSMUST00000229439.2
|
Tcf20
|
transcription factor 20 |
| chr14_-_16968099 | 1.18 |
ENSMUST00000181562.8
|
Gm3488
|
predicted gene, 3488 |
| chr7_+_36397426 | 1.16 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr13_-_92268156 | 1.16 |
ENSMUST00000151408.9
ENSMUST00000216219.2 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr11_+_117545618 | 1.15 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr14_-_24054352 | 1.14 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr19_-_34856853 | 1.13 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr13_+_119565669 | 1.13 |
ENSMUST00000173627.8
ENSMUST00000126957.9 ENSMUST00000174691.8 |
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr17_+_26934617 | 1.12 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr7_+_101859542 | 1.12 |
ENSMUST00000140631.2
ENSMUST00000120879.8 ENSMUST00000146996.8 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chrX_-_73067514 | 1.11 |
ENSMUST00000033769.15
ENSMUST00000114352.8 ENSMUST00000068286.12 ENSMUST00000114360.10 ENSMUST00000114354.10 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr18_+_82929037 | 1.11 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr7_+_128858730 | 1.10 |
ENSMUST00000094018.6
ENSMUST00000205896.2 |
Plpp4
|
phospholipid phosphatase 4 |
| chr18_+_36414122 | 1.09 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr6_+_107506678 | 1.08 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr3_-_107366868 | 1.08 |
ENSMUST00000009617.10
ENSMUST00000238670.2 |
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr11_+_108811168 | 1.08 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr1_+_59521583 | 1.07 |
ENSMUST00000114246.4
|
Fzd7
|
frizzled class receptor 7 |
| chr13_-_60324755 | 1.07 |
ENSMUST00000223933.2
|
Gas1
|
growth arrest specific 1 |
| chr15_+_39061612 | 1.06 |
ENSMUST00000082054.12
ENSMUST00000227243.2 ENSMUST00000042917.10 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr13_+_43070127 | 1.06 |
ENSMUST00000239286.2
|
Phactr1
|
phosphatase and actin regulator 1 |
| chrX_-_73067351 | 1.05 |
ENSMUST00000114353.10
ENSMUST00000101458.9 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_-_157240138 | 1.05 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr7_+_141047298 | 1.04 |
ENSMUST00000106000.10
ENSMUST00000209892.2 ENSMUST00000177840.9 |
Cd151
|
CD151 antigen |
| chr7_+_141047416 | 1.04 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr2_-_80411578 | 1.02 |
ENSMUST00000028386.12
|
Nckap1
|
NCK-associated protein 1 |
| chr13_-_31743316 | 1.01 |
ENSMUST00000170573.2
|
A530084C06Rik
|
RIKEN cDNA A530084C06 gene |
| chr7_+_141055135 | 1.01 |
ENSMUST00000026585.14
|
Tspan4
|
tetraspanin 4 |
| chr4_+_11156411 | 1.01 |
ENSMUST00000029865.4
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr15_+_76227695 | 1.01 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr14_-_18135010 | 1.00 |
ENSMUST00000165466.8
|
2610042L04Rik
|
RIKEN cDNA 2610042L04 gene |
| chr5_-_24806960 | 0.99 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr5_-_76452365 | 0.99 |
ENSMUST00000075159.5
|
Clock
|
circadian locomotor output cycles kaput |
| chr13_+_73775001 | 0.98 |
ENSMUST00000022104.9
|
Tert
|
telomerase reverse transcriptase |
| chr17_-_85995680 | 0.98 |
ENSMUST00000024947.8
ENSMUST00000163568.4 |
Six2
|
sine oculis-related homeobox 2 |
| chr4_-_45530330 | 0.98 |
ENSMUST00000061986.12
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr15_+_34838195 | 0.97 |
ENSMUST00000228725.2
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr3_-_104127676 | 0.96 |
ENSMUST00000064371.14
|
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr2_+_84489890 | 0.96 |
ENSMUST00000133437.3
|
Btbd18
|
BTB (POZ) domain containing 18 |
| chr14_-_18897750 | 0.96 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr14_-_18659699 | 0.96 |
ENSMUST00000170480.8
|
Gm3002
|
predicted gene 3002 |
| chr18_+_82929451 | 0.94 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr12_+_95658987 | 0.94 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr8_+_108020132 | 0.93 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr5_-_37981919 | 0.93 |
ENSMUST00000063116.10
|
Msx1
|
msh homeobox 1 |
| chr6_+_72333209 | 0.92 |
ENSMUST00000206531.2
|
Tmem150a
|
transmembrane protein 150A |
| chr9_-_43117052 | 0.91 |
ENSMUST00000176636.5
|
Pou2f3
|
POU domain, class 2, transcription factor 3 |
| chr11_+_94218810 | 0.90 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_-_166838018 | 0.90 |
ENSMUST00000049412.12
|
Stau1
|
staufen double-stranded RNA binding protein 1 |
| chr14_-_24054273 | 0.89 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_+_82216800 | 0.89 |
ENSMUST00000222298.2
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_104127419 | 0.88 |
ENSMUST00000121198.8
ENSMUST00000122303.2 |
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr5_+_135197228 | 0.88 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr11_+_70431063 | 0.88 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr3_+_37117676 | 0.88 |
ENSMUST00000144629.8
|
Adad1
|
adenosine deaminase domain containing 1 (testis specific) |
| chr7_+_141055350 | 0.88 |
ENSMUST00000138092.8
ENSMUST00000146305.8 |
Tspan4
|
tetraspanin 4 |
| chr3_+_37117779 | 0.88 |
ENSMUST00000029274.14
|
Adad1
|
adenosine deaminase domain containing 1 (testis specific) |
| chr6_-_83433357 | 0.87 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr11_+_59197746 | 0.87 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr2_-_73605684 | 0.87 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chrX_+_7439839 | 0.87 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr2_-_166838182 | 0.85 |
ENSMUST00000109238.9
ENSMUST00000109235.8 ENSMUST00000109236.9 |
Stau1
|
staufen double-stranded RNA binding protein 1 |
| chr7_-_132724344 | 0.85 |
ENSMUST00000167218.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_+_77821626 | 0.84 |
ENSMUST00000093995.10
ENSMUST00000000646.14 |
Sez6
|
seizure related gene 6 |
| chr10_-_127502541 | 0.84 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 2.2 | 15.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 1.4 | 4.1 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 1.3 | 6.5 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.2 | 7.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 1.1 | 6.7 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 1.0 | 4.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.9 | 2.7 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.9 | 3.6 | GO:0015755 | fructose transport(GO:0015755) |
| 0.9 | 2.6 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.6 | 6.7 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.5 | 1.6 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.5 | 4.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.5 | 1.6 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.5 | 1.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 2.9 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.5 | 2.8 | GO:2000054 | regulation of mismatch repair(GO:0032423) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 1.8 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.4 | 2.7 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.4 | 1.7 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.4 | 1.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.4 | 1.5 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.4 | 1.1 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.3 | 0.7 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.3 | 1.0 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.3 | 1.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 1.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 5.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 0.9 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.3 | 2.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 0.9 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.3 | 4.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 0.8 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.3 | 1.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 1.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.3 | 1.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 2.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 1.0 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.2 | 1.5 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.2 | 4.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 1.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 0.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.2 | 2.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 1.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 0.8 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 0.6 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.6 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 1.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.5 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.2 | 0.9 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 1.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 1.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 1.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 0.6 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 | 0.9 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.2 | 1.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 1.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 1.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 1.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 1.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 1.9 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 0.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 1.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 3.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 2.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.0 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.5 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.7 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.5 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 1.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 1.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 1.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.7 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 3.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.1 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 1.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 2.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 2.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 3.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.0 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.7 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.3 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 2.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 4.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.4 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.2 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 3.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 1.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.3 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 1.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.7 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.7 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.3 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 1.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 1.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.3 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.1 | GO:0061038 | estradiol secretion(GO:0035938) uterus morphogenesis(GO:0061038) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.7 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.2 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.5 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.6 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 2.0 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.3 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 2.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.7 | 3.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 1.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.0 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 1.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 3.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 15.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 17.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 2.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 2.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 4.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 4.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.0 | 1.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 5.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 3.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 7.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 1.2 | 3.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) uniporter activity(GO:0015292) |
| 1.0 | 2.9 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.8 | 4.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.8 | 6.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.7 | 2.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.7 | 2.1 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.6 | 3.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 1.6 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.5 | 1.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.4 | 1.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.4 | 2.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 4.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 6.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.4 | 1.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 1.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 2.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 2.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 4.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 3.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.7 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.2 | 1.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 2.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 7.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.8 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 1.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 3.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 4.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.9 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 1.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 4.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 4.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 3.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 23.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 4.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 3.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 4.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 3.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 4.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 7.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 7.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 4.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 3.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 6.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 4.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 4.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |