Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Z-value: 1.84
Transcription factors associated with Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
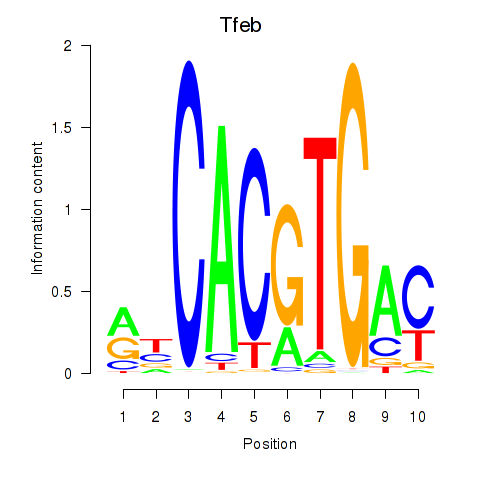
Tfeb
|
ENSMUSG00000023990.19 | transcription factor EB |
|
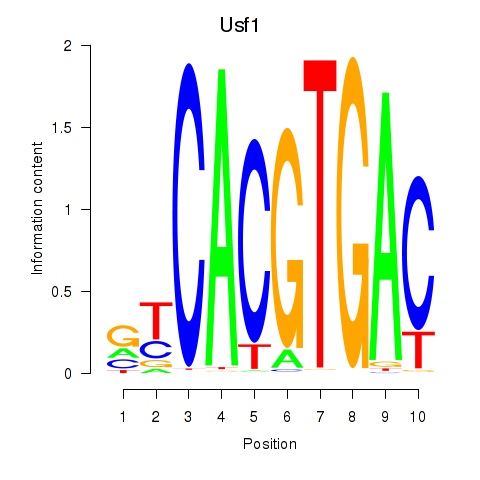
Usf1
|
ENSMUSG00000026641.14 | upstream transcription factor 1 |
|
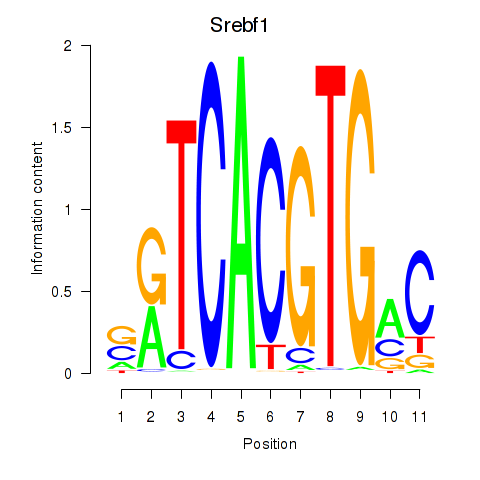
Srebf1
|
ENSMUSG00000020538.16 | sterol regulatory element binding transcription factor 1 |
|
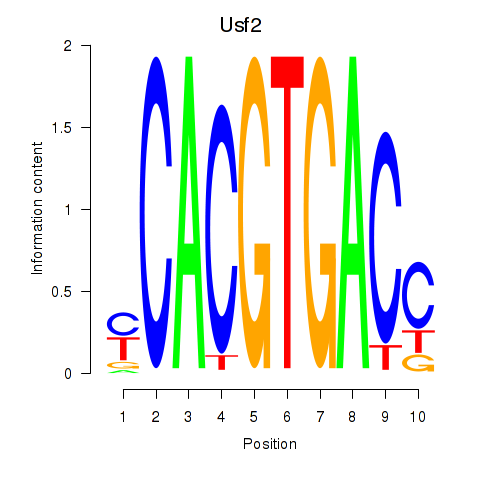
Usf2
|
ENSMUSG00000058239.14 | upstream transcription factor 2 |
|
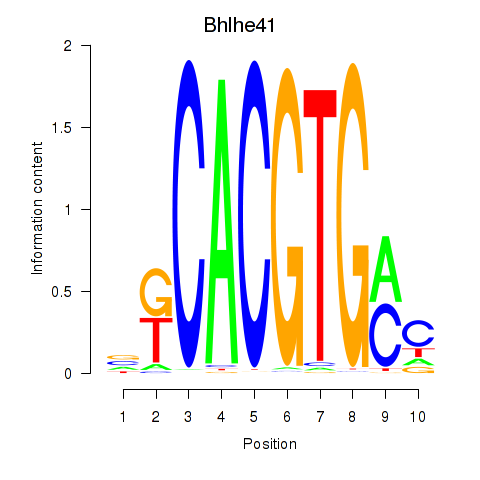
Bhlhe41
|
ENSMUSG00000030256.12 | basic helix-loop-helix family, member e41 |
|
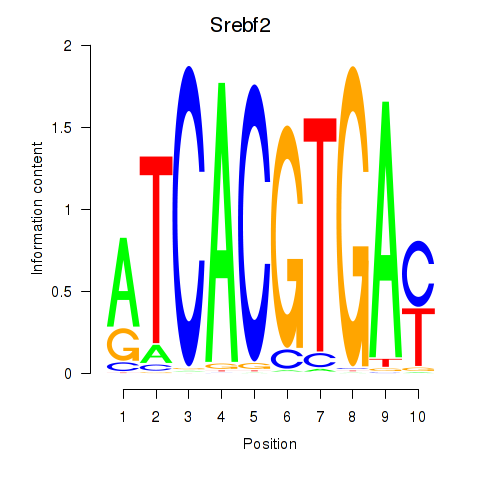
Srebf2
|
ENSMUSG00000022463.9 | sterol regulatory element binding factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Usf2 | mm39_v1_chr7_-_30656167_30656228 | 0.68 | 5.2e-06 | Click! |
| Srebf2 | mm39_v1_chr15_+_82031382_82031476 | 0.66 | 1.3e-05 | Click! |
| Srebf1 | mm39_v1_chr11_-_60111391_60111458 | -0.52 | 1.0e-03 | Click! |
| Bhlhe41 | mm39_v1_chr6_-_145811274_145811284 | 0.48 | 2.8e-03 | Click! |
| Usf1 | mm39_v1_chr1_+_171238873_171238906 | 0.48 | 2.9e-03 | Click! |
| Tfeb | mm39_v1_chr17_+_48047955_48048033 | 0.44 | 7.7e-03 | Click! |
Activity profile of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
Sorted Z-values of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 66.0 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 7.3 | 21.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 5.7 | 39.8 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 4.2 | 12.6 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 3.9 | 11.6 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 3.7 | 11.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 3.5 | 34.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 3.4 | 10.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 3.2 | 16.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 3.1 | 18.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 2.9 | 17.3 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 2.9 | 14.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 2.5 | 7.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 2.4 | 16.9 | GO:0010288 | response to lead ion(GO:0010288) |
| 2.4 | 14.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 2.3 | 11.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 2.2 | 6.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.0 | 12.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.0 | 7.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.9 | 11.2 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 1.8 | 7.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 1.8 | 21.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.7 | 5.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.5 | 19.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.5 | 4.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 1.5 | 7.5 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 1.5 | 28.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 1.5 | 5.8 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 1.5 | 11.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.4 | 5.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 1.4 | 11.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 1.4 | 2.8 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.4 | 4.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 1.4 | 9.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.4 | 15.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.3 | 3.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 1.3 | 35.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 1.3 | 80.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 1.3 | 8.8 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 1.2 | 6.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 1.2 | 10.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 1.2 | 4.8 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 1.2 | 3.6 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 1.2 | 13.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.2 | 18.8 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 1.1 | 11.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.1 | 2.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.1 | 7.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.1 | 6.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.0 | 4.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.0 | 6.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.0 | 16.0 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 1.0 | 10.9 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 1.0 | 4.9 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.0 | 3.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.0 | 3.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 1.0 | 3.9 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 1.0 | 1.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.0 | 6.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.9 | 31.6 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.9 | 3.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.9 | 3.6 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.9 | 21.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.9 | 0.9 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.9 | 4.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.8 | 3.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 7.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.8 | 3.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.8 | 21.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.8 | 3.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.8 | 8.7 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.8 | 2.4 | GO:0045212 | acetylcholine metabolic process(GO:0008291) neurotransmitter receptor biosynthetic process(GO:0045212) acetate ester metabolic process(GO:1900619) |
| 0.8 | 10.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.8 | 6.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.8 | 10.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.8 | 1.5 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.7 | 2.9 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.7 | 12.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.7 | 14.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.7 | 2.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.7 | 8.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.7 | 2.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 12.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.6 | 4.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.6 | 2.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.6 | 2.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.6 | 5.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.6 | 2.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.6 | 1.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 2.9 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.6 | 1.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.6 | 1.7 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 | 2.8 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.6 | 5.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 6.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.5 | 5.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.5 | 3.2 | GO:0030421 | defecation(GO:0030421) |
| 0.5 | 13.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.5 | 3.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 2.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.5 | 4.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.5 | 4.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.5 | 2.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.5 | 2.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.5 | 4.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 1.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 2.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.5 | 1.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.5 | 1.9 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.5 | 2.8 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 5.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.5 | 2.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 11.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.5 | 0.5 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.5 | 1.4 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.5 | 4.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.5 | 4.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 1.8 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.5 | 2.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 3.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 0.9 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.4 | 0.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 2.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.4 | 8.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.4 | 1.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 9.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.4 | 5.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.4 | 1.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.7 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.4 | 2.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.4 | 24.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.4 | 2.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 11.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 1.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.4 | 1.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.4 | 0.8 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.4 | 6.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.4 | 3.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.4 | 4.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.4 | 1.5 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.4 | 1.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.4 | 0.7 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.4 | 1.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 1.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 4.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 5.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 1.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 0.3 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.3 | 39.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.3 | 8.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.3 | 2.9 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.3 | 2.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 2.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 18.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.3 | 0.6 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.3 | 5.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.3 | 5.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 3.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 12.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.3 | 0.6 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 | 1.4 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 | 0.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.3 | 8.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 2.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 1.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 2.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 1.9 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 0.8 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.3 | 1.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.3 | 0.8 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.3 | 7.9 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.3 | 3.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 1.3 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.3 | 1.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 1.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 1.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 2.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 3.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 0.7 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.2 | 19.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 3.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.2 | 7.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 1.2 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 19.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.2 | 0.9 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 2.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.7 | GO:0060540 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.2 | 2.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 1.8 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 2.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 1.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 1.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.9 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 2.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.2 | 1.0 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 3.5 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.2 | 2.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 0.6 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 2.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.2 | 1.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 5.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 1.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 7.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 6.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 7.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 3.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.2 | 2.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 1.8 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 5.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 5.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 0.7 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 24.2 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.2 | 1.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 6.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.2 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 0.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 2.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 18.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 4.0 | GO:0071548 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 14.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 1.3 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.2 | 19.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.2 | 4.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 3.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 7.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 4.5 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 4.3 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 2.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 3.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 2.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 3.0 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 4.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 3.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.9 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 28.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 9.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 | 1.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.6 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 3.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 3.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.8 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 1.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.2 | GO:0072318 | protein targeting to vacuole involved in autophagy(GO:0071211) clathrin coat disassembly(GO:0072318) |
| 0.1 | 1.9 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 9.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.5 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.1 | 1.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.8 | GO:0014857 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.9 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 2.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 2.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.4 | GO:0061418 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 1.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 7.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.5 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 2.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.5 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 4.8 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 4.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 2.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 4.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 2.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.5 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 0.5 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 4.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.2 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 4.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 3.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 1.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.6 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 1.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 6.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 3.1 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.3 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.5 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 1.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 2.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.7 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 1.0 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.3 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.3 | GO:0043153 | photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.4 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.5 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 61.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 6.6 | 19.9 | GO:1990879 | CST complex(GO:1990879) |
| 4.2 | 12.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 4.2 | 12.6 | GO:0097144 | BAX complex(GO:0097144) |
| 3.9 | 19.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 3.5 | 24.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 3.2 | 13.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.7 | 8.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 2.5 | 10.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 2.3 | 9.0 | GO:0035101 | FACT complex(GO:0035101) |
| 2.1 | 16.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 2.1 | 10.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 2.0 | 16.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.0 | 10.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.9 | 31.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.8 | 12.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.8 | 17.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.7 | 1.7 | GO:0044094 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 1.6 | 21.1 | GO:0042555 | MCM complex(GO:0042555) |
| 1.5 | 9.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.5 | 4.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.3 | 6.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.3 | 12.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.2 | 14.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.2 | 11.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 1.1 | 6.7 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.1 | 6.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 1.1 | 2.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.0 | 15.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.0 | 3.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.9 | 15.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 6.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.8 | 11.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.8 | 7.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.8 | 60.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.8 | 6.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.7 | 3.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.7 | 3.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 2.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.7 | 13.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.7 | 20.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.6 | 5.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.6 | 4.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 6.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 4.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.5 | 1.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.5 | 18.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.5 | 4.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 2.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 2.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.4 | 3.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 5.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 3.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.4 | 2.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 5.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.4 | 1.6 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.4 | 1.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.4 | 2.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 2.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.4 | 3.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 3.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 15.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 2.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.3 | 4.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 4.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 2.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 18.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 4.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 1.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 8.9 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 5.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 1.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 8.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 19.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 3.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 16.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 6.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 1.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 3.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 25.7 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 1.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 4.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 3.2 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.2 | 25.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 19.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 2.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 61.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 2.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 9.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 2.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 0.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 27.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 3.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 4.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 6.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 9.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 3.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 3.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 6.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 4.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 3.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 63.2 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.1 | 16.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 12.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 2.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 31.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 8.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 4.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 10.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 4.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 2.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 14.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 8.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 7.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.9 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.5 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 7.3 | 21.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 4.0 | 60.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 4.0 | 12.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 4.0 | 19.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 3.7 | 18.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 3.5 | 10.6 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 3.5 | 31.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 2.6 | 7.9 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 2.4 | 12.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.9 | 59.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 1.8 | 12.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.8 | 9.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.8 | 5.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.8 | 17.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.8 | 7.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.7 | 5.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 1.7 | 10.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.7 | 8.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.6 | 4.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.5 | 7.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.5 | 2.9 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 1.5 | 8.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.5 | 11.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.5 | 8.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.4 | 21.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.4 | 5.7 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.4 | 4.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 1.4 | 4.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.4 | 4.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 1.3 | 12.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.2 | 4.7 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.1 | 4.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.1 | 4.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.1 | 7.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.0 | 3.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 1.0 | 3.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.0 | 17.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.0 | 14.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.0 | 45.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 1.0 | 4.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 29.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.9 | 12.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.9 | 4.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.9 | 5.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.9 | 11.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.9 | 6.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.8 | 50.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.8 | 9.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.8 | 10.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.8 | 21.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.8 | 3.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.8 | 8.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.8 | 3.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.8 | 87.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.8 | 12.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.8 | 2.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.8 | 3.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.8 | 2.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.8 | 5.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.8 | 1.5 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.7 | 2.2 | GO:0003726 | left-handed Z-DNA binding(GO:0003692) double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.7 | 10.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.7 | 2.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.7 | 6.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 6.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 5.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 2.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.6 | 4.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 4.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 2.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.5 | 1.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.5 | 2.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 5.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 3.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.5 | 3.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.5 | 26.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.5 | 1.8 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.5 | 1.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.4 | 4.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.3 | GO:0061749 | forked DNA-dependent helicase activity(GO:0061749) |
| 0.4 | 24.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 1.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.4 | 4.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 2.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 4.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 31.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 21.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.4 | 11.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.4 | 2.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 10.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 11.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 3.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 4.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 3.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 5.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 13.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.4 | 1.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 8.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 6.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 3.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.4 | 1.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 7.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.3 | 3.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 6.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 5.9 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.3 | 1.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 2.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 2.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 1.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 3.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 2.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 3.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 3.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 19.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.3 | 7.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 0.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.3 | 3.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 2.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 7.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 4.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 9.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 3.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 2.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 14.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 3.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 5.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 14.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 1.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 2.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 2.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 7.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.7 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 6.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 1.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 3.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 8.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 64.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 3.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 3.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 0.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 0.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 0.6 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 3.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 9.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 6.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 16.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 30.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.7 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 5.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 4.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.8 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 8.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 4.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 38.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 14.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 5.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.3 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 2.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 6.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 9.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 5.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.8 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 13.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 4.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 12.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:1902121 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.0 | 1.1 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.5 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 6.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 25.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.0 | 16.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 47.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.8 | 17.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.7 | 26.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 3.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.7 | 3.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.5 | 22.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 18.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.4 | 14.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 10.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 4.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 10.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 34.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 10.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 9.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 13.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 8.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 2.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 7.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 6.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 15.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 5.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 9.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 5.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 7.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 11.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 4.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 4.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 9.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 10.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 5.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 9.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 7.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 6.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 4.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 24.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.3 | 32.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.3 | 21.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 40.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.9 | 14.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.8 | 23.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.7 | 55.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.7 | 8.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.7 | 4.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.6 | 22.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.6 | 36.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 12.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.5 | 84.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.5 | 19.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.5 | 3.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.5 | 14.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 2.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 24.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 10.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 2.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.4 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 6.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 5.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 19.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 24.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 4.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 18.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 4.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 20.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 4.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 19.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 6.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 1.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 11.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 4.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 3.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 14.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 4.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 4.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 2.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 4.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 12.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 10.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 14.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 6.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 13.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |