Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
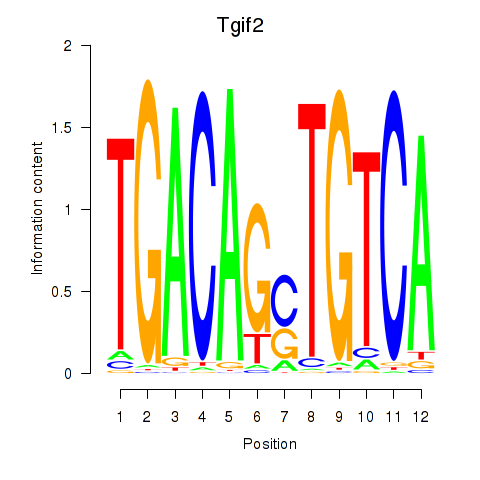
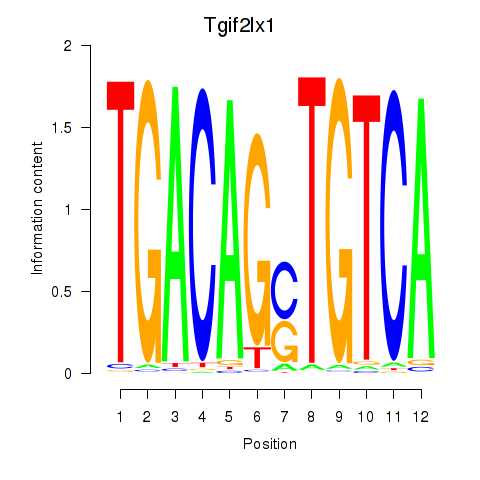
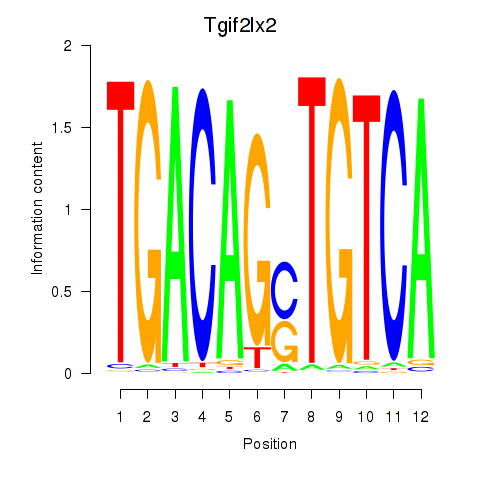
Results for Tgif2_Tgif2lx1_Tgif2lx2
Z-value: 0.56
Transcription factors associated with Tgif2_Tgif2lx1_Tgif2lx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tgif2
|
ENSMUSG00000062175.14 | TGFB-induced factor homeobox 2 |
|
Tgif2lx1
|
ENSMUSG00000100133.2 | TGFB-induced factor homeobox 2-like, X-linked 1 |
|
Tgif2lx2
|
ENSMUSG00000100194.2 | TGFB-induced factor homeobox 2-like, X-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tgif2 | mm39_v1_chr2_+_156681991_156682062 | 0.34 | 4.5e-02 | Click! |
Activity profile of Tgif2_Tgif2lx1_Tgif2lx2 motif
Sorted Z-values of Tgif2_Tgif2lx1_Tgif2lx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_54849268 | 4.49 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr9_+_108437485 | 2.86 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr4_+_114945905 | 2.84 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr7_+_19144950 | 2.69 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr11_-_86999481 | 2.58 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr13_+_108452866 | 2.39 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chrX_+_8137881 | 2.30 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr15_-_54953819 | 2.30 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr4_-_116228921 | 2.19 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr11_+_58808716 | 2.08 |
ENSMUST00000069941.13
|
Btnl10
|
butyrophilin-like 10 |
| chr2_-_84652890 | 2.06 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr9_+_64188857 | 2.02 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr11_+_117877118 | 1.88 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr2_-_129139125 | 1.83 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr11_+_87000032 | 1.61 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_-_127689890 | 1.59 |
ENSMUST00000057198.9
ENSMUST00000199273.2 |
Fam241a
|
family with sequence similarity 241, member A |
| chr13_+_108452930 | 1.55 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr4_-_87724533 | 1.48 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_58808830 | 1.35 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr7_-_83384711 | 1.23 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr11_+_115768323 | 1.19 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr7_-_142223662 | 1.19 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr10_-_87982732 | 1.16 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr4_-_155013002 | 1.15 |
ENSMUST00000152687.8
ENSMUST00000137803.8 ENSMUST00000145296.2 |
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr6_-_121058551 | 1.13 |
ENSMUST00000077159.8
ENSMUST00000207889.2 ENSMUST00000238957.2 ENSMUST00000238920.2 ENSMUST00000238968.2 |
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr8_-_71292295 | 1.09 |
ENSMUST00000212405.2
ENSMUST00000002989.11 |
Arrdc2
|
arrestin domain containing 2 |
| chr18_-_62313019 | 1.06 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr3_+_107784543 | 1.03 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr9_-_107956474 | 1.00 |
ENSMUST00000162355.8
ENSMUST00000047746.13 ENSMUST00000174504.8 ENSMUST00000178267.8 ENSMUST00000160649.8 |
Rnf123
|
ring finger protein 123 |
| chr4_-_87724512 | 0.99 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_79784365 | 0.92 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr16_+_49620883 | 0.90 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr6_+_112436466 | 0.86 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr2_+_110427643 | 0.80 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr6_-_83302890 | 0.76 |
ENSMUST00000204472.2
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr8_+_85753452 | 0.74 |
ENSMUST00000047281.10
|
Trir
|
telomerase RNA component interacting RNase |
| chr11_+_101207743 | 0.71 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr5_+_136145485 | 0.70 |
ENSMUST00000111127.8
ENSMUST00000041366.14 ENSMUST00000111129.2 |
Polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr7_+_28140352 | 0.70 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr9_+_48273333 | 0.69 |
ENSMUST00000093853.5
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr10_+_80662490 | 0.68 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr5_+_31212165 | 0.67 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr5_+_31212110 | 0.64 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr1_+_36346824 | 0.63 |
ENSMUST00000142319.8
ENSMUST00000097778.9 ENSMUST00000115031.8 ENSMUST00000115032.8 ENSMUST00000137906.2 ENSMUST00000115029.2 |
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr15_+_44320918 | 0.62 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr17_-_56019571 | 0.62 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr7_+_28140450 | 0.60 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr5_+_35146727 | 0.58 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_+_106768714 | 0.58 |
ENSMUST00000072753.13
ENSMUST00000097934.10 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr16_+_33614378 | 0.57 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr14_-_8123295 | 0.55 |
ENSMUST00000225775.2
ENSMUST00000224877.2 ENSMUST00000090591.4 |
Il3ra
|
interleukin 3 receptor, alpha chain |
| chr4_+_106768662 | 0.55 |
ENSMUST00000030367.15
ENSMUST00000149926.8 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr1_-_169359015 | 0.53 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr9_-_107956453 | 0.53 |
ENSMUST00000162516.3
|
Rnf123
|
ring finger protein 123 |
| chr1_-_74040723 | 0.50 |
ENSMUST00000190389.7
|
Tns1
|
tensin 1 |
| chr17_-_46513499 | 0.50 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr5_-_114911548 | 0.49 |
ENSMUST00000178440.8
ENSMUST00000043283.14 ENSMUST00000112185.9 ENSMUST00000155908.8 |
Git2
|
GIT ArfGAP 2 |
| chr8_-_123338157 | 0.48 |
ENSMUST00000015171.11
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr8_-_123187406 | 0.47 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr8_-_70892204 | 0.41 |
ENSMUST00000076615.6
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr8_-_123338126 | 0.41 |
ENSMUST00000212319.2
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr8_+_123338357 | 0.39 |
ENSMUST00000015157.10
|
Trappc2l
|
trafficking protein particle complex 2-like |
| chr5_-_114911432 | 0.39 |
ENSMUST00000112183.8
|
Git2
|
GIT ArfGAP 2 |
| chr7_+_48438751 | 0.38 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr1_+_128079543 | 0.38 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr1_-_182929025 | 0.37 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr5_-_114911509 | 0.37 |
ENSMUST00000086564.11
|
Git2
|
GIT ArfGAP 2 |
| chr2_+_26209755 | 0.36 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr2_+_157298836 | 0.36 |
ENSMUST00000109529.2
|
Src
|
Rous sarcoma oncogene |
| chr17_+_87332978 | 0.36 |
ENSMUST00000024959.10
|
Cript
|
cysteine-rich PDZ-binding protein |
| chr8_+_104977493 | 0.36 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr10_+_87982854 | 0.33 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr14_+_32935983 | 0.33 |
ENSMUST00000165792.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr3_+_96577447 | 0.33 |
ENSMUST00000048427.9
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr9_+_44045859 | 0.33 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr6_+_88442391 | 0.33 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr13_-_63006176 | 0.33 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr16_+_20536856 | 0.32 |
ENSMUST00000231392.2
ENSMUST00000161038.2 |
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_138058139 | 0.32 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr7_+_30411634 | 0.32 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr5_-_36739724 | 0.31 |
ENSMUST00000031094.15
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chrX_+_93410718 | 0.29 |
ENSMUST00000113898.8
|
Apoo
|
apolipoprotein O |
| chr9_+_50679528 | 0.28 |
ENSMUST00000042391.13
|
Fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr15_-_85695855 | 0.28 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr2_-_121337120 | 0.27 |
ENSMUST00000089926.6
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr2_-_174314672 | 0.27 |
ENSMUST00000117442.8
ENSMUST00000141100.2 ENSMUST00000120822.2 |
Prelid3b
|
PRELI domain containing 3B |
| chr2_-_34716199 | 0.27 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr5_-_124233812 | 0.26 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr8_+_95055829 | 0.26 |
ENSMUST00000034218.5
ENSMUST00000212134.2 |
Slc12a3
|
solute carrier family 12, member 3 |
| chr2_-_34716083 | 0.26 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chrX_+_48552803 | 0.25 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr17_+_29309942 | 0.25 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr7_+_106692427 | 0.25 |
ENSMUST00000210568.4
|
Olfr17
|
olfactory receptor 17 |
| chr17_+_30845909 | 0.24 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr5_-_134581235 | 0.24 |
ENSMUST00000036999.10
ENSMUST00000100647.7 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr18_+_53309388 | 0.24 |
ENSMUST00000037850.7
|
Snx2
|
sorting nexin 2 |
| chr2_-_120561983 | 0.24 |
ENSMUST00000110701.8
ENSMUST00000110700.2 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr1_-_106980033 | 0.24 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr10_+_39245746 | 0.24 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr7_+_45683122 | 0.24 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr2_-_174314741 | 0.24 |
ENSMUST00000016401.15
|
Prelid3b
|
PRELI domain containing 3B |
| chr17_-_25105277 | 0.23 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr3_+_101917455 | 0.23 |
ENSMUST00000066187.6
ENSMUST00000198675.2 |
Nhlh2
|
nescient helix loop helix 2 |
| chrX_+_93410754 | 0.22 |
ENSMUST00000113897.9
ENSMUST00000113896.8 ENSMUST00000113895.2 |
Apoo
|
apolipoprotein O |
| chr11_+_76792977 | 0.22 |
ENSMUST00000102495.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_34760960 | 0.22 |
ENSMUST00000028225.12
|
Psmd5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr6_+_142359099 | 0.22 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr17_+_46513666 | 0.22 |
ENSMUST00000087031.7
|
Xpo5
|
exportin 5 |
| chr3_+_138047536 | 0.22 |
ENSMUST00000199673.6
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr10_-_120037464 | 0.21 |
ENSMUST00000020448.11
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr9_+_86625694 | 0.21 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr11_-_114892706 | 0.18 |
ENSMUST00000092464.10
|
Cd300c2
|
CD300C molecule 2 |
| chr6_+_68657317 | 0.18 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr6_-_69792108 | 0.18 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr11_+_78717398 | 0.18 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr5_+_145282064 | 0.17 |
ENSMUST00000079268.9
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr19_-_5660057 | 0.16 |
ENSMUST00000236229.2
ENSMUST00000235701.2 ENSMUST00000236264.2 |
Kat5
|
K(lysine) acetyltransferase 5 |
| chr7_-_30523191 | 0.16 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr15_-_85695291 | 0.16 |
ENSMUST00000125947.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr4_+_132903646 | 0.16 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr5_+_139392142 | 0.16 |
ENSMUST00000052176.9
|
C130050O18Rik
|
RIKEN cDNA C130050O18 gene |
| chr3_-_63391300 | 0.16 |
ENSMUST00000192926.2
|
Strit1
|
small transmembrane regulator of ion transport 1 |
| chr19_-_46950948 | 0.16 |
ENSMUST00000236924.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr15_-_85695810 | 0.16 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr6_-_68681962 | 0.16 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr1_+_118409769 | 0.15 |
ENSMUST00000191823.6
|
Clasp1
|
CLIP associating protein 1 |
| chr14_-_51168260 | 0.15 |
ENSMUST00000160538.2
ENSMUST00000162957.8 ENSMUST00000161166.8 ENSMUST00000160835.9 ENSMUST00000049312.14 |
Pip4p1
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1 |
| chr5_-_103359117 | 0.15 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr11_+_31950452 | 0.14 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr5_+_125080504 | 0.14 |
ENSMUST00000197746.2
|
Rflna
|
refilin A |
| chr2_+_32611067 | 0.14 |
ENSMUST00000074248.11
|
Sh2d3c
|
SH2 domain containing 3C |
| chr17_+_25105617 | 0.14 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr10_+_87982916 | 0.14 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr2_+_3337194 | 0.13 |
ENSMUST00000115089.2
ENSMUST00000228935.2 |
Acbd7
|
acyl-Coenzyme A binding domain containing 7 |
| chr12_-_115459678 | 0.13 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr3_+_105811712 | 0.13 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr11_+_68393845 | 0.12 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr10_+_92996686 | 0.12 |
ENSMUST00000069965.9
|
Cdk17
|
cyclin-dependent kinase 17 |
| chr1_+_135980488 | 0.12 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr5_-_94763020 | 0.12 |
ENSMUST00000180178.2
|
Pramel43
|
PRAME like 43 |
| chr5_-_136275407 | 0.12 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr2_-_7400690 | 0.11 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr3_-_92336712 | 0.11 |
ENSMUST00000192538.2
ENSMUST00000047300.8 |
Gm9774
|
predicted pseudogene 9774 |
| chr1_+_135980639 | 0.11 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr2_-_120801186 | 0.11 |
ENSMUST00000028728.6
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr11_+_30904390 | 0.10 |
ENSMUST00000020551.13
ENSMUST00000117883.2 |
Asb3
|
ankyrin repeat and SOCS box-containing 3 |
| chr1_+_135980508 | 0.10 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr5_-_100307130 | 0.10 |
ENSMUST00000139520.3
|
Tmem150c
|
transmembrane protein 150C |
| chr11_-_55042529 | 0.10 |
ENSMUST00000020502.9
ENSMUST00000069816.6 |
Slc36a3
|
solute carrier family 36 (proton/amino acid symporter), member 3 |
| chr14_-_69945022 | 0.10 |
ENSMUST00000118374.8
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr16_+_6887689 | 0.10 |
ENSMUST00000229741.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_8122634 | 0.10 |
ENSMUST00000223589.2
|
Il3ra
|
interleukin 3 receptor, alpha chain |
| chr4_+_146098479 | 0.09 |
ENSMUST00000131932.2
|
Zfp600
|
zinc finger protein 600 |
| chr1_+_184936306 | 0.09 |
ENSMUST00000194740.6
ENSMUST00000069652.8 |
Rab3gap2
|
RAB3 GTPase activating protein subunit 2 |
| chr12_-_103660916 | 0.09 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr6_+_67816777 | 0.09 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chr11_+_85243970 | 0.09 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr1_-_3741721 | 0.08 |
ENSMUST00000070533.5
|
Xkr4
|
X-linked Kx blood group related 4 |
| chr15_+_82031382 | 0.08 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr5_-_94075731 | 0.08 |
ENSMUST00000179824.2
|
Pramel36
|
PRAME like 36 |
| chr19_-_33369655 | 0.08 |
ENSMUST00000163093.2
|
Rnls
|
renalase, FAD-dependent amine oxidase |
| chr6_+_83303052 | 0.08 |
ENSMUST00000038658.15
|
Mob1a
|
MOB kinase activator 1A |
| chr6_+_123679629 | 0.07 |
ENSMUST00000172391.4
|
Vmn2r23
|
vomeronasal 2, receptor 23 |
| chr8_+_26339646 | 0.07 |
ENSMUST00000098858.11
|
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr17_+_34258411 | 0.07 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr15_-_12549350 | 0.07 |
ENSMUST00000190929.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr15_-_94302139 | 0.07 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr13_+_41267783 | 0.07 |
ENSMUST00000124093.3
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr2_-_7400780 | 0.07 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr19_-_11852453 | 0.07 |
ENSMUST00000213954.2
ENSMUST00000217617.2 |
Olfr1419
|
olfactory receptor 1419 |
| chr3_+_101917392 | 0.07 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr4_-_82778066 | 0.07 |
ENSMUST00000107239.8
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr7_-_18532425 | 0.07 |
ENSMUST00000004657.14
ENSMUST00000182853.2 |
Psg19
|
pregnancy specific glycoprotein 19 |
| chr4_+_136150835 | 0.07 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr10_-_81332147 | 0.07 |
ENSMUST00000118498.8
|
Ncln
|
nicalin |
| chr4_-_116024788 | 0.07 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr16_-_20972750 | 0.07 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr5_-_131567526 | 0.06 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr17_+_14168635 | 0.06 |
ENSMUST00000088809.6
|
Gm7168
|
predicted gene 7168 |
| chr6_-_88851579 | 0.06 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr6_+_83302998 | 0.06 |
ENSMUST00000055261.11
|
Mob1a
|
MOB kinase activator 1A |
| chr18_+_37847792 | 0.06 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr7_+_120682472 | 0.06 |
ENSMUST00000171880.3
ENSMUST00000047025.15 ENSMUST00000170106.2 ENSMUST00000168311.8 |
Otoa
|
otoancorin |
| chr6_+_116528102 | 0.05 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chr6_-_142750192 | 0.05 |
ENSMUST00000111768.3
|
Sult6b2
|
sulfotransferase family 6B, member 2 |
| chr7_-_42228012 | 0.05 |
ENSMUST00000179470.2
|
Gm21028
|
predicted gene, 21028 |
| chr10_+_5589210 | 0.05 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr17_+_30843328 | 0.04 |
ENSMUST00000170651.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr7_-_28879879 | 0.04 |
ENSMUST00000209034.2
ENSMUST00000182328.8 ENSMUST00000186182.2 ENSMUST00000059642.17 |
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr6_-_69741999 | 0.04 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr2_+_130509530 | 0.04 |
ENSMUST00000103193.5
|
Itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr5_+_109478028 | 0.04 |
ENSMUST00000165180.3
ENSMUST00000232912.2 |
Vmn2r16
|
vomeronasal 2, receptor 16 |
| chr2_-_165210622 | 0.04 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr8_+_13755884 | 0.04 |
ENSMUST00000166277.5
ENSMUST00000214337.2 |
Cfap97d2
|
CFAP97 domain containing 2 |
| chr1_-_163552693 | 0.04 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr14_+_53628092 | 0.04 |
ENSMUST00000103636.4
|
Trav7-2
|
T cell receptor alpha variable 7-2 |
| chr7_-_106315603 | 0.04 |
ENSMUST00000216868.2
|
Olfr695
|
olfactory receptor 695 |
| chr7_+_106462392 | 0.03 |
ENSMUST00000080925.7
|
Olfr704
|
olfactory receptor 704 |
| chr18_-_77134939 | 0.03 |
ENSMUST00000137354.8
ENSMUST00000137498.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr1_-_130867810 | 0.03 |
ENSMUST00000112465.2
ENSMUST00000187410.7 ENSMUST00000187916.7 |
Il19
|
interleukin 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tgif2_Tgif2lx1_Tgif2lx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.5 | 2.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 1.3 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.4 | 2.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 2.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.4 | 1.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 | 1.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 0.7 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.6 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 1.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 1.2 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.2 | 1.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 2.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.4 | GO:2000554 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.1 | 0.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 2.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 2.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.9 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 1.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 2.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.9 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 1.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 3.8 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.5 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.1 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 1.6 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 2.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 2.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 2.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 2.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 0.7 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.2 | 2.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 1.9 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.8 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 1.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 5.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 4.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |