Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Thra
Z-value: 0.50
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSMUSG00000058756.14 | thyroid hormone receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | mm39_v1_chr11_+_98632696_98632720 | 0.74 | 3.1e-07 | Click! |
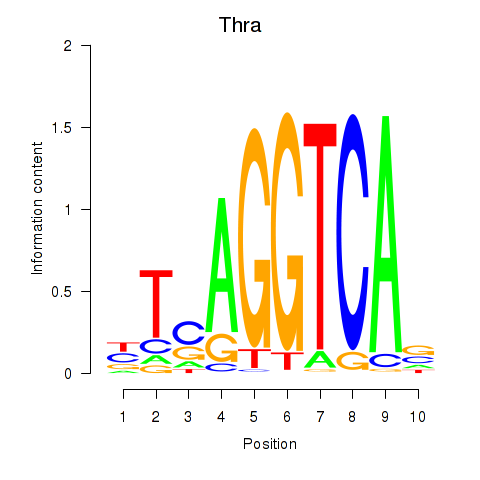
Activity profile of Thra motif
Sorted Z-values of Thra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_7834057 | 3.84 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr9_+_110848339 | 3.72 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr1_-_84817022 | 2.71 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_+_131028861 | 2.67 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr11_-_70146156 | 2.53 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr4_+_44300876 | 2.40 |
ENSMUST00000045607.12
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr13_+_95833359 | 1.79 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chrX_-_142610371 | 1.79 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr15_+_73595012 | 1.62 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr15_+_73594965 | 1.62 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr7_+_141995545 | 1.57 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr16_-_19801781 | 1.55 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr19_-_46033353 | 1.53 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr5_+_110987839 | 1.34 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr10_-_62363217 | 1.32 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chrX_-_56384089 | 1.29 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr6_+_86605146 | 1.26 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr7_+_28140352 | 1.22 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr1_-_134006847 | 1.20 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr17_-_71575584 | 1.19 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr11_-_40624200 | 1.18 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr5_-_110987604 | 1.17 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr11_+_97340962 | 1.17 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr1_-_170886924 | 1.16 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr6_+_120813162 | 1.14 |
ENSMUST00000203584.3
ENSMUST00000203037.3 |
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr1_+_172327569 | 1.12 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr4_-_156340276 | 1.07 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr6_+_120813199 | 1.05 |
ENSMUST00000009256.4
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr7_+_19144950 | 1.02 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr1_-_170133901 | 0.99 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr11_+_69856222 | 0.99 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr6_+_113508636 | 0.98 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr15_-_78413816 | 0.93 |
ENSMUST00000023075.9
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr17_+_48571298 | 0.92 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr4_+_94627513 | 0.92 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr8_+_72889073 | 0.91 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr4_-_62278761 | 0.91 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr13_+_20274708 | 0.90 |
ENSMUST00000072519.7
|
Elmo1
|
engulfment and cell motility 1 |
| chr4_-_126096376 | 0.89 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr7_+_28140450 | 0.89 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr4_-_41275091 | 0.89 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr4_-_137493785 | 0.89 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr1_-_84817000 | 0.87 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr3_+_96736774 | 0.85 |
ENSMUST00000138014.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr3_+_96079642 | 0.84 |
ENSMUST00000076372.5
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr3_+_96736600 | 0.84 |
ENSMUST00000135031.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr8_+_121264161 | 0.84 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr4_-_156340713 | 0.84 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr3_+_95071617 | 0.83 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr7_+_3339077 | 0.83 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_-_163822336 | 0.83 |
ENSMUST00000097493.10
ENSMUST00000045876.8 |
BC055324
|
cDNA sequence BC055324 |
| chr16_-_18904240 | 0.82 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr15_-_78413780 | 0.82 |
ENSMUST00000229185.2
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr13_+_55357585 | 0.81 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr10_-_87982732 | 0.80 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr7_+_3339059 | 0.78 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr4_-_140501507 | 0.76 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr6_+_136509922 | 0.75 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_+_32250043 | 0.73 |
ENSMUST00000115140.2
ENSMUST00000104893.10 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr4_-_49597837 | 0.72 |
ENSMUST00000042750.3
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr4_+_33310306 | 0.71 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr19_-_6835538 | 0.70 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr14_-_73563212 | 0.70 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr5_-_110987441 | 0.70 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr1_+_152683627 | 0.67 |
ENSMUST00000027754.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr5_-_74226394 | 0.67 |
ENSMUST00000145016.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr7_-_100164007 | 0.67 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr11_+_31822211 | 0.65 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_+_44409775 | 0.64 |
ENSMUST00000019333.10
|
Rnf145
|
ring finger protein 145 |
| chr4_-_144921567 | 0.62 |
ENSMUST00000036579.14
|
Vps13d
|
vacuolar protein sorting 13D |
| chr7_+_26958150 | 0.61 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr11_+_72332167 | 0.61 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_+_4936824 | 0.59 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr4_-_135000109 | 0.58 |
ENSMUST00000037099.9
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
| chr18_+_60907668 | 0.57 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr1_+_152683568 | 0.56 |
ENSMUST00000190323.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr15_+_65682066 | 0.56 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr14_+_25459690 | 0.55 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr8_-_106434565 | 0.54 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr15_+_34083450 | 0.53 |
ENSMUST00000163333.9
ENSMUST00000170050.8 ENSMUST00000170553.8 |
Mtdh
|
metadherin |
| chr18_+_60907698 | 0.53 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr11_-_29497819 | 0.52 |
ENSMUST00000102844.4
|
Rps27a
|
ribosomal protein S27A |
| chr7_+_18817767 | 0.52 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr7_+_79674562 | 0.50 |
ENSMUST00000164056.9
ENSMUST00000166250.8 |
Zfp710
|
zinc finger protein 710 |
| chr1_+_134121170 | 0.48 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr11_+_75541324 | 0.48 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
| chr3_+_122522592 | 0.47 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr1_-_84817976 | 0.44 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr16_-_23339329 | 0.44 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr4_-_49597425 | 0.44 |
ENSMUST00000150664.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr10_-_7556881 | 0.43 |
ENSMUST00000159977.2
ENSMUST00000162682.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr14_-_55344004 | 0.43 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr7_+_12656217 | 0.43 |
ENSMUST00000108539.8
ENSMUST00000004554.14 ENSMUST00000147435.8 ENSMUST00000137329.4 |
Rps5
|
ribosomal protein S5 |
| chr15_-_82796308 | 0.42 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr4_-_126096551 | 0.42 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr15_+_99600149 | 0.42 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr2_+_112096154 | 0.42 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr10_-_79940168 | 0.42 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr11_-_85030761 | 0.41 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr5_+_33021042 | 0.40 |
ENSMUST00000149350.8
ENSMUST00000118698.8 ENSMUST00000150130.8 ENSMUST00000049780.13 ENSMUST00000087897.11 ENSMUST00000119705.8 ENSMUST00000125574.8 |
Depdc5
|
DEP domain containing 5 |
| chr5_+_34527230 | 0.40 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr4_-_58912678 | 0.40 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr17_+_64203017 | 0.39 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr3_+_113824181 | 0.39 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr1_-_152262425 | 0.39 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chrX_-_12026594 | 0.38 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chrX_+_98086187 | 0.37 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr14_+_25459630 | 0.36 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr14_+_79718604 | 0.35 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr3_-_94693780 | 0.35 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr14_-_79718890 | 0.34 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr4_-_141327146 | 0.33 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr5_-_123804745 | 0.33 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr9_+_107784065 | 0.32 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr15_+_78314251 | 0.32 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr16_+_91282121 | 0.31 |
ENSMUST00000023689.11
ENSMUST00000117748.8 |
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chr16_+_32249713 | 0.31 |
ENSMUST00000115137.8
ENSMUST00000079791.11 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr3_+_95067759 | 0.31 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_83741657 | 0.31 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr15_-_98063441 | 0.31 |
ENSMUST00000123922.2
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chrX_+_168468186 | 0.30 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr11_+_83741689 | 0.30 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr4_-_41048124 | 0.29 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr16_+_72460029 | 0.29 |
ENSMUST00000023600.8
|
Robo1
|
roundabout guidance receptor 1 |
| chr1_-_134162231 | 0.29 |
ENSMUST00000169927.2
|
Adora1
|
adenosine A1 receptor |
| chr18_+_82932747 | 0.29 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chrX_+_100492684 | 0.28 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr11_+_115366470 | 0.28 |
ENSMUST00000035240.7
|
Armc7
|
armadillo repeat containing 7 |
| chr18_+_35987733 | 0.28 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr1_-_152262339 | 0.28 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr17_-_32607859 | 0.28 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr2_+_22785534 | 0.28 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr10_+_126814542 | 0.28 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr11_+_70655035 | 0.28 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chr12_-_40087393 | 0.27 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chrX_-_52357897 | 0.27 |
ENSMUST00000114838.8
|
Fam122b
|
family with sequence similarity 122, member B |
| chr5_-_134975773 | 0.27 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr18_+_9957906 | 0.27 |
ENSMUST00000025137.9
|
Thoc1
|
THO complex 1 |
| chr2_-_34716199 | 0.26 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr18_-_64794338 | 0.26 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr12_+_117480099 | 0.26 |
ENSMUST00000109691.4
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr19_+_31846154 | 0.26 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr12_+_78908466 | 0.25 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr11_+_120348919 | 0.25 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr17_+_23898223 | 0.25 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr2_-_65069383 | 0.25 |
ENSMUST00000155916.8
ENSMUST00000156643.2 |
Cobll1
|
Cobl-like 1 |
| chr4_+_131600918 | 0.25 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr4_-_141391406 | 0.24 |
ENSMUST00000084203.11
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr6_+_83114020 | 0.24 |
ENSMUST00000121093.8
|
Rtkn
|
rhotekin |
| chr19_-_7688628 | 0.23 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr10_+_7556948 | 0.23 |
ENSMUST00000165952.9
|
Lats1
|
large tumor suppressor |
| chr3_+_31150982 | 0.23 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr3_-_95214443 | 0.22 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr16_-_31094095 | 0.22 |
ENSMUST00000060188.14
|
Ppp1r2
|
protein phosphatase 1, regulatory inhibitor subunit 2 |
| chr16_+_91282183 | 0.21 |
ENSMUST00000129878.2
|
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chr7_-_126574959 | 0.21 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr11_+_40624763 | 0.21 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr1_+_159871943 | 0.21 |
ENSMUST00000163892.8
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chrX_+_135567124 | 0.21 |
ENSMUST00000060904.11
ENSMUST00000113100.2 ENSMUST00000128040.2 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr13_+_44883270 | 0.20 |
ENSMUST00000172830.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_97206542 | 0.19 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr8_+_72021567 | 0.19 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr15_-_98063493 | 0.18 |
ENSMUST00000143400.8
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr8_+_72021510 | 0.18 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr6_+_83114086 | 0.18 |
ENSMUST00000087938.11
|
Rtkn
|
rhotekin |
| chrX_+_143902497 | 0.18 |
ENSMUST00000096301.5
|
Rtl4
|
retrotransposon Gag like 4 |
| chr18_+_65158873 | 0.18 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr8_+_73197718 | 0.18 |
ENSMUST00000064853.13
ENSMUST00000121902.2 |
1700030K09Rik
|
RIKEN cDNA 1700030K09 gene |
| chr15_-_74624811 | 0.18 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr2_-_119308094 | 0.18 |
ENSMUST00000110808.2
ENSMUST00000049920.14 |
Ino80
|
INO80 complex subunit |
| chr10_+_87982854 | 0.17 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr4_-_139695337 | 0.16 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr12_+_102435383 | 0.16 |
ENSMUST00000179218.9
|
Golga5
|
golgi autoantigen, golgin subfamily a, 5 |
| chr19_-_46315543 | 0.16 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr10_+_87982916 | 0.15 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr12_+_102435425 | 0.15 |
ENSMUST00000021609.10
|
Golga5
|
golgi autoantigen, golgin subfamily a, 5 |
| chr3_+_86131970 | 0.15 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr3_+_122523219 | 0.15 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr3_+_108191398 | 0.14 |
ENSMUST00000135636.6
ENSMUST00000102632.7 |
Sort1
|
sortilin 1 |
| chr7_+_24990596 | 0.13 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr4_+_126042250 | 0.13 |
ENSMUST00000106150.3
|
Eva1b
|
eva-1 homolog B (C. elegans) |
| chr12_-_98703664 | 0.13 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr1_-_181985663 | 0.13 |
ENSMUST00000169123.4
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr7_+_100122192 | 0.13 |
ENSMUST00000032958.14
ENSMUST00000107059.2 |
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr7_-_125799570 | 0.13 |
ENSMUST00000009344.16
|
Xpo6
|
exportin 6 |
| chr4_-_128856213 | 0.13 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr6_+_115337899 | 0.12 |
ENSMUST00000171644.8
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr18_+_42408418 | 0.12 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr15_+_99600475 | 0.11 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr5_+_53747556 | 0.11 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr18_+_35987791 | 0.11 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr10_+_84412490 | 0.11 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr1_+_131838220 | 0.11 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr7_+_126575510 | 0.10 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr11_+_97697328 | 0.10 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr4_-_109333866 | 0.10 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr16_-_19060440 | 0.10 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
| chr10_+_93476903 | 0.09 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr2_-_79959178 | 0.09 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_+_80614795 | 0.09 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
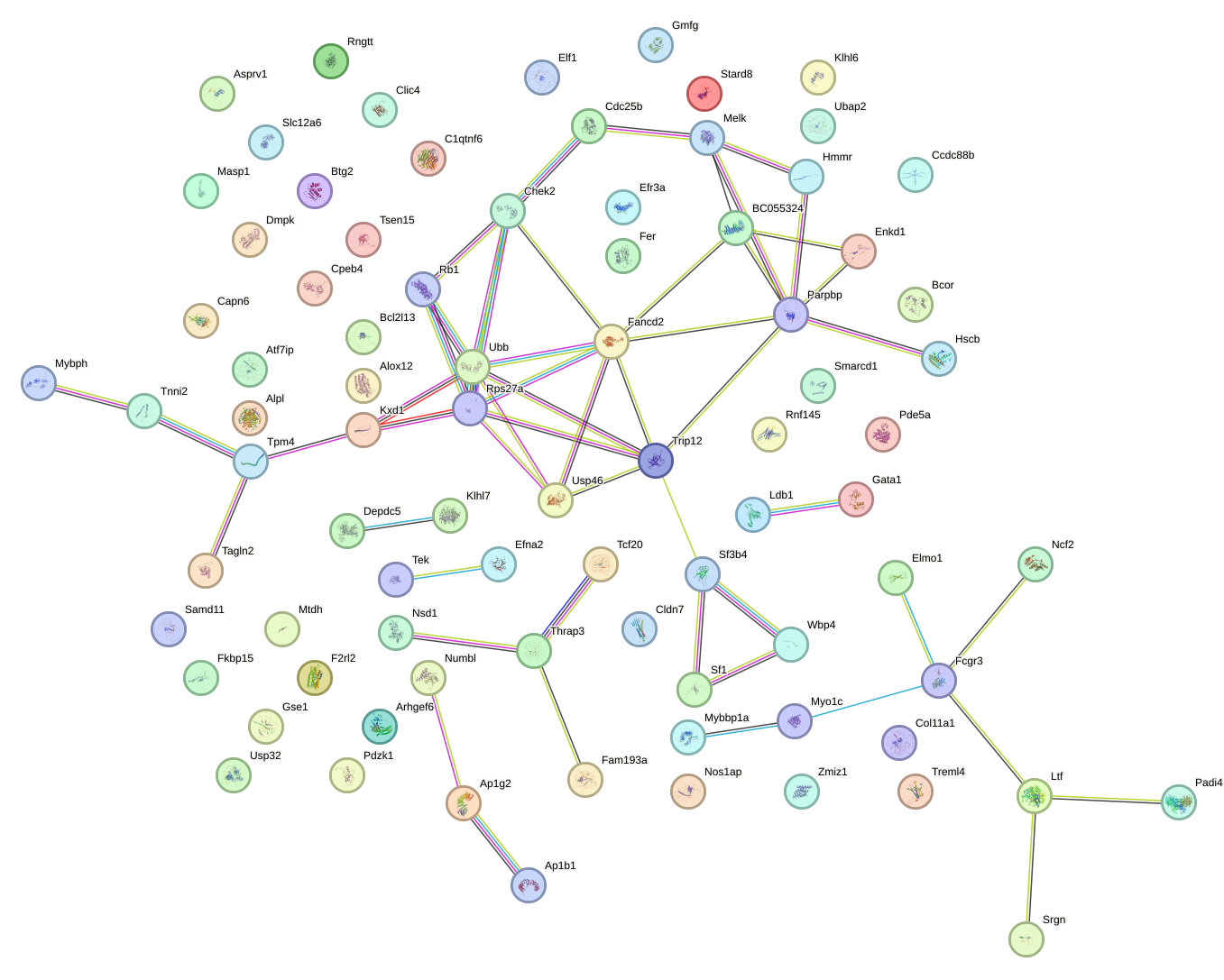
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.9 | 3.7 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.7 | 2.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.7 | 4.0 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.5 | 1.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.5 | 2.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 1.3 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.4 | 1.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.3 | 1.3 | GO:0033370 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.3 | 2.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.2 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.7 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 3.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.7 | GO:0061215 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.2 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 0.8 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 1.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.3 | GO:0042323 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.7 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 1.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 2.3 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.0 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 2.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 1.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.4 | 1.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 3.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 0.5 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.2 | 1.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 6.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 1.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 5.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 4.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 3.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |