Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Thrb
Z-value: 1.26
Transcription factors associated with Thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thrb
|
ENSMUSG00000021779.20 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thrb | mm39_v1_chr14_-_4488167_4488189 | 0.42 | 1.1e-02 | Click! |
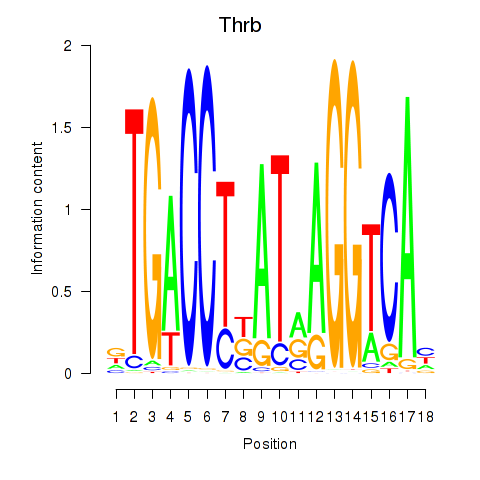
Activity profile of Thrb motif
Sorted Z-values of Thrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105775224 | 23.66 |
ENSMUST00000093222.13
ENSMUST00000093223.5 |
Ces3a
|
carboxylesterase 3A |
| chr3_+_14928561 | 23.47 |
ENSMUST00000029076.6
|
Car3
|
carbonic anhydrase 3 |
| chr7_+_25597045 | 16.20 |
ENSMUST00000072438.13
ENSMUST00000005477.6 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chr2_+_58457370 | 15.04 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr7_-_105249308 | 9.95 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr10_+_87357782 | 9.08 |
ENSMUST00000219813.2
|
Pah
|
phenylalanine hydroxylase |
| chr17_+_37253802 | 8.94 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr15_-_76193955 | 6.88 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr2_+_118998235 | 6.50 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr10_+_87357657 | 6.47 |
ENSMUST00000020241.17
|
Pah
|
phenylalanine hydroxylase |
| chr14_-_61597843 | 5.37 |
ENSMUST00000022494.10
|
Ebpl
|
emopamil binding protein-like |
| chr14_+_14475188 | 4.72 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr11_-_113600346 | 4.66 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr12_+_108300599 | 4.41 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr9_-_107546166 | 4.32 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr4_+_41760454 | 4.17 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr9_-_107546195 | 4.08 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr2_-_25359752 | 3.83 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr17_+_37253916 | 3.40 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr11_-_113600838 | 3.40 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr9_+_44584523 | 3.29 |
ENSMUST00000034609.11
ENSMUST00000071219.12 |
Treh
|
trehalase (brush-border membrane glycoprotein) |
| chr10_+_77522403 | 2.91 |
ENSMUST00000092366.4
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr2_-_25360043 | 2.62 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr1_-_52539395 | 2.20 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr4_+_116078787 | 2.12 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr4_+_20042045 | 2.12 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr17_+_57297280 | 1.82 |
ENSMUST00000002735.9
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr13_+_34923589 | 1.61 |
ENSMUST00000221037.2
|
Fam50b
|
family with sequence similarity 50, member B |
| chr9_+_102503815 | 1.56 |
ENSMUST00000038673.14
ENSMUST00000186693.2 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chr5_+_95021656 | 1.45 |
ENSMUST00000180076.2
|
Gm3183
|
predicted gene 3183 |
| chr5_-_74692327 | 1.38 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr9_+_102503476 | 1.38 |
ENSMUST00000190279.7
ENSMUST00000188398.7 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chrX_-_77627388 | 1.35 |
ENSMUST00000177904.8
|
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr6_+_114435480 | 1.29 |
ENSMUST00000160780.2
|
Hrh1
|
histamine receptor H1 |
| chr5_+_94365864 | 1.23 |
ENSMUST00000179743.3
|
Pramel38
|
PRAME like 38 |
| chr3_-_79749949 | 1.21 |
ENSMUST00000029568.7
|
Tmem144
|
transmembrane protein 144 |
| chr11_-_58516867 | 1.20 |
ENSMUST00000203173.2
ENSMUST00000070804.3 |
Olfr323
|
olfactory receptor 323 |
| chr3_-_36529753 | 0.93 |
ENSMUST00000199478.2
|
Anxa5
|
annexin A5 |
| chr19_-_46314945 | 0.89 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr4_+_116078830 | 0.89 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr16_-_16962256 | 0.86 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr16_-_28571820 | 0.86 |
ENSMUST00000232352.2
|
Fgf12
|
fibroblast growth factor 12 |
| chr11_+_70350436 | 0.81 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr2_+_152804405 | 0.80 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chrX_-_77627486 | 0.78 |
ENSMUST00000114025.8
ENSMUST00000134602.8 ENSMUST00000114024.9 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr14_+_53315909 | 0.76 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8 |
| chr16_+_8498618 | 0.72 |
ENSMUST00000201722.2
|
Gm5767
|
predicted gene 5767 |
| chr1_-_24626492 | 0.68 |
ENSMUST00000051344.6
ENSMUST00000115244.9 |
Col19a1
|
collagen, type XIX, alpha 1 |
| chr17_-_28908757 | 0.64 |
ENSMUST00000233398.2
|
Slc26a8
|
solute carrier family 26, member 8 |
| chr10_-_18662526 | 0.56 |
ENSMUST00000216654.2
|
Gm4922
|
predicted gene 4922 |
| chr11_+_70350252 | 0.53 |
ENSMUST00000108563.9
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr17_-_14223966 | 0.50 |
ENSMUST00000189454.3
|
Gm7356
|
predicted gene 7356 |
| chr8_-_120228434 | 0.49 |
ENSMUST00000133821.2
ENSMUST00000036748.15 |
Slc38a8
|
solute carrier family 38, member 8 |
| chr14_+_54000594 | 0.40 |
ENSMUST00000103589.6
|
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr11_+_109376432 | 0.38 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr9_-_102503180 | 0.38 |
ENSMUST00000216281.2
ENSMUST00000093791.10 |
Cep63
|
centrosomal protein 63 |
| chr13_-_92667321 | 0.36 |
ENSMUST00000022217.9
|
Zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr5_+_35971697 | 0.36 |
ENSMUST00000130233.8
|
Ablim2
|
actin-binding LIM protein 2 |
| chr11_+_49094292 | 0.34 |
ENSMUST00000150284.8
ENSMUST00000109197.8 ENSMUST00000151228.2 |
Zfp62
|
zinc finger protein 62 |
| chr13_-_59892731 | 0.30 |
ENSMUST00000180139.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr7_+_27222678 | 0.30 |
ENSMUST00000108353.9
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr16_-_16962279 | 0.29 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr8_+_112262729 | 0.27 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr13_+_38041910 | 0.20 |
ENSMUST00000138110.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr6_-_121450547 | 0.06 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr14_+_54713703 | 0.05 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr1_-_83385911 | 0.01 |
ENSMUST00000160953.8
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0009095 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 2.8 | 8.4 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 2.3 | 16.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.4 | 10.0 | GO:0060332 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.1 | 6.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.7 | 8.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 3.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.5 | 2.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 23.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 4.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 1.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 4.7 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 4.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 0.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.9 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 15.0 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 6.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.1 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.8 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 2.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 6.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 5.4 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 2.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 8.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 6.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 6.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 10.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 46.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 8.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.5 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 2.5 | 15.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.1 | 8.4 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.8 | 23.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.7 | 10.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.6 | 6.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.0 | 4.2 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.8 | 6.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 16.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.3 | 3.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 5.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 6.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 8.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 23.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 2.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 4.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 4.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.2 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 10.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 15.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 6.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 4.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 6.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 8.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 15.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |