Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
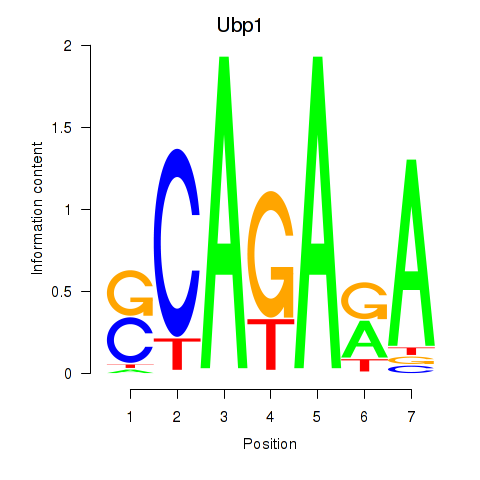
Results for Ubp1
Z-value: 0.86
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSMUSG00000009741.16 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | mm39_v1_chr9_+_113760376_113760722 | -0.24 | 1.7e-01 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13357892 | 3.62 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr3_-_98800524 | 3.37 |
ENSMUST00000029464.9
|
Hao2
|
hydroxyacid oxidase 2 |
| chr16_+_48692976 | 3.04 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr1_-_172722589 | 3.01 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr9_+_110867807 | 2.94 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr7_-_26638802 | 2.44 |
ENSMUST00000170227.3
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr5_+_90920294 | 2.37 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr15_+_89218601 | 2.28 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr3_+_10077608 | 2.26 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr2_-_28453374 | 2.21 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr14_-_56339915 | 2.14 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr11_+_101258368 | 2.03 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr14_-_60324265 | 1.98 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr5_+_90920353 | 1.96 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr6_+_121613177 | 1.94 |
ENSMUST00000032203.9
|
A2m
|
alpha-2-macroglobulin |
| chr11_+_104468107 | 1.92 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_-_106446928 | 1.88 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chrX_-_142610371 | 1.85 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr10_-_111833138 | 1.85 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr8_-_4155758 | 1.83 |
ENSMUST00000138439.2
ENSMUST00000145007.8 |
Cd209f
|
CD209f antigen |
| chr2_-_26800581 | 1.82 |
ENSMUST00000015920.12
ENSMUST00000139815.2 ENSMUST00000102899.10 |
Med22
|
mediator complex subunit 22 |
| chr2_+_148640705 | 1.79 |
ENSMUST00000028931.10
ENSMUST00000109947.2 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr11_-_101998648 | 1.75 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr6_+_30639217 | 1.73 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_-_40976413 | 1.69 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr17_-_7639378 | 1.67 |
ENSMUST00000231397.2
|
Gm49630
|
predicted gene, 49630 |
| chr17_+_28426752 | 1.66 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr12_+_113115632 | 1.65 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr1_+_172327812 | 1.65 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr1_-_45542442 | 1.65 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chrX_+_8137620 | 1.59 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_+_69945157 | 1.58 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chrX_-_8011952 | 1.56 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr10_+_127612243 | 1.56 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chrX_+_8137881 | 1.52 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr10_+_127595639 | 1.47 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr17_+_32755525 | 1.45 |
ENSMUST00000169591.8
ENSMUST00000003416.15 |
Cyp4f16
|
cytochrome P450, family 4, subfamily f, polypeptide 16 |
| chr13_-_55676334 | 1.45 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chrX_-_8011918 | 1.44 |
ENSMUST00000115619.8
ENSMUST00000115617.10 ENSMUST00000040010.10 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_-_68713748 | 1.42 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr10_-_128384971 | 1.40 |
ENSMUST00000176906.2
|
Rpl41
|
ribosomal protein L41 |
| chr11_+_117740111 | 1.37 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr9_+_51124983 | 1.37 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr6_-_41423004 | 1.35 |
ENSMUST00000095999.7
|
Gm10334
|
predicted gene 10334 |
| chr17_+_37180437 | 1.34 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr8_+_57964921 | 1.33 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr6_-_83513222 | 1.31 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_3848050 | 1.31 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr11_+_70529944 | 1.30 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr6_+_41523664 | 1.30 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr10_-_128384994 | 1.29 |
ENSMUST00000177163.8
ENSMUST00000176683.8 ENSMUST00000176010.8 |
Rpl41
|
ribosomal protein L41 |
| chr7_-_126817475 | 1.28 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr12_+_85733415 | 1.28 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr11_-_102815910 | 1.25 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr7_-_140596811 | 1.25 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr7_+_78563513 | 1.23 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr17_+_29309942 | 1.22 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chrX_+_10351360 | 1.20 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr8_+_117822593 | 1.20 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr7_+_92469100 | 1.19 |
ENSMUST00000107180.8
ENSMUST00000107179.2 |
Rab30
|
RAB30, member RAS oncogene family |
| chr14_-_30645503 | 1.16 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_-_70149254 | 1.16 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr6_+_67838100 | 1.16 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr3_-_10273628 | 1.16 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr11_+_72851989 | 1.15 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr11_+_115768323 | 1.15 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr15_+_78128990 | 1.15 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr4_+_114914880 | 1.15 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr15_+_79400597 | 1.15 |
ENSMUST00000010974.9
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr6_-_142418801 | 1.13 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr18_-_33597060 | 1.12 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr6_+_38895902 | 1.10 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr12_-_103956176 | 1.10 |
ENSMUST00000151709.3
ENSMUST00000176246.3 ENSMUST00000074693.13 ENSMUST00000120251.9 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr13_-_19803928 | 1.09 |
ENSMUST00000221014.2
ENSMUST00000002885.8 ENSMUST00000220944.2 |
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr7_-_126303887 | 1.08 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_-_75060345 | 1.08 |
ENSMUST00000055619.5
|
Hic1
|
hypermethylated in cancer 1 |
| chr1_-_170755109 | 1.08 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr2_-_158071124 | 1.07 |
ENSMUST00000103121.10
ENSMUST00000169335.8 ENSMUST00000046944.6 |
D630003M21Rik
|
RIKEN cDNA D630003M21 gene |
| chr5_-_134258435 | 1.07 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr19_+_12577317 | 1.07 |
ENSMUST00000181868.8
ENSMUST00000092931.7 |
Gm4952
|
predicted gene 4952 |
| chr6_+_34361153 | 1.04 |
ENSMUST00000038383.14
ENSMUST00000115051.8 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr7_+_78563150 | 1.02 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr7_+_142030744 | 1.02 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr14_-_25903524 | 1.01 |
ENSMUST00000052286.16
|
Plac9a
|
placenta specific 9a |
| chr14_-_30645711 | 1.00 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr2_-_164198427 | 1.00 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr19_-_10181243 | 1.00 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr11_+_121593219 | 0.99 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr19_-_10581622 | 0.98 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr15_-_54953819 | 0.98 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr2_+_24235300 | 0.97 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr13_-_34119816 | 0.96 |
ENSMUST00000168350.8
ENSMUST00000167260.8 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr10_-_81335966 | 0.95 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr8_+_57964956 | 0.95 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr2_-_32314017 | 0.95 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr7_-_111758241 | 0.95 |
ENSMUST00000033036.7
|
Dkk3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr18_+_62086122 | 0.95 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr16_-_18884431 | 0.94 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr7_+_44866095 | 0.93 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr1_+_135656885 | 0.93 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr17_+_84933872 | 0.93 |
ENSMUST00000025101.10
|
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr2_-_151815307 | 0.93 |
ENSMUST00000109863.2
|
Fam110a
|
family with sequence similarity 110, member A |
| chr2_-_33777874 | 0.93 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr2_+_120331784 | 0.92 |
ENSMUST00000151342.3
|
Capn3
|
calpain 3 |
| chr8_-_125296435 | 0.92 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr15_+_78129040 | 0.92 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr6_-_70051586 | 0.91 |
ENSMUST00000103377.3
|
Igkv6-32
|
immunoglobulin kappa variable 6-32 |
| chr1_+_164624200 | 0.90 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr11_-_69496655 | 0.90 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr9_+_46179899 | 0.89 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr10_+_127851031 | 0.88 |
ENSMUST00000178041.8
ENSMUST00000026461.8 |
Prim1
|
DNA primase, p49 subunit |
| chr8_+_4184652 | 0.87 |
ENSMUST00000130372.3
|
Cd209g
|
CD209g antigen |
| chr7_-_43309563 | 0.87 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr3_-_82053068 | 0.87 |
ENSMUST00000048976.8
|
Gucy1a1
|
guanylate cyclase 1, soluble, alpha 1 |
| chr4_-_156340276 | 0.87 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr12_-_113589576 | 0.86 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr2_+_90912710 | 0.85 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_-_50401305 | 0.85 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr12_+_99850756 | 0.85 |
ENSMUST00000153627.8
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr15_+_101310283 | 0.84 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr5_+_31454787 | 0.83 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr6_-_113411718 | 0.83 |
ENSMUST00000113091.8
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr6_-_42350188 | 0.81 |
ENSMUST00000073387.5
ENSMUST00000204357.2 |
Epha1
|
Eph receptor A1 |
| chr4_+_130643260 | 0.81 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr6_-_125471666 | 0.81 |
ENSMUST00000032492.9
|
Cd9
|
CD9 antigen |
| chr2_-_32321116 | 0.81 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr7_-_4815542 | 0.80 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr6_+_38918327 | 0.80 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr6_-_83513184 | 0.80 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_-_126916487 | 0.80 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr10_+_21868114 | 0.79 |
ENSMUST00000150089.8
ENSMUST00000100036.10 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_39396902 | 0.79 |
ENSMUST00000122996.8
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr17_+_34524841 | 0.79 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr6_+_112250719 | 0.79 |
ENSMUST00000032376.6
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr4_+_130253925 | 0.78 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr2_+_84966569 | 0.78 |
ENSMUST00000057019.9
|
Aplnr
|
apelin receptor |
| chr8_-_4309257 | 0.76 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr1_-_171061902 | 0.76 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr9_-_71075939 | 0.76 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr8_-_106665060 | 0.75 |
ENSMUST00000034369.10
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr11_-_53959790 | 0.75 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr7_-_99002204 | 0.74 |
ENSMUST00000208292.2
ENSMUST00000207989.2 ENSMUST00000208749.2 ENSMUST00000169437.9 ENSMUST00000208119.2 ENSMUST00000207849.2 |
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr3_+_20011201 | 0.74 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr11_+_121593582 | 0.74 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr6_+_41515152 | 0.73 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr7_-_99002430 | 0.73 |
ENSMUST00000094154.6
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr2_+_26800757 | 0.73 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr18_-_35855383 | 0.73 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr17_-_28705055 | 0.72 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr6_-_69792108 | 0.72 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr1_-_143879877 | 0.72 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr11_-_69586884 | 0.72 |
ENSMUST00000180587.8
|
Tnfsfm13
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 |
| chr12_-_113802603 | 0.72 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr6_-_5496261 | 0.72 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr18_+_36661198 | 0.71 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr5_-_113957362 | 0.71 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr13_-_55661240 | 0.70 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr6_+_35229589 | 0.70 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_-_109986763 | 0.70 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr5_-_31250817 | 0.70 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chrX_-_47551990 | 0.69 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr7_-_30023320 | 0.69 |
ENSMUST00000098586.5
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr14_-_104081827 | 0.69 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr11_-_72441054 | 0.68 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr10_-_76562002 | 0.68 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr2_+_103800553 | 0.68 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr6_+_142244145 | 0.68 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr7_-_19411866 | 0.68 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr2_+_103800459 | 0.67 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr18_-_36859732 | 0.67 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr14_-_56181993 | 0.67 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr2_+_103799873 | 0.66 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr7_-_44888532 | 0.66 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr15_+_82225380 | 0.66 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2 |
| chr6_-_69753317 | 0.66 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr1_+_171041539 | 0.66 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_34170401 | 0.65 |
ENSMUST00000087543.5
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr4_+_140427799 | 0.65 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr11_-_79421397 | 0.65 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chrX_+_138701544 | 0.65 |
ENSMUST00000054889.4
|
Cldn2
|
claudin 2 |
| chr6_-_83010402 | 0.64 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr6_-_48549594 | 0.64 |
ENSMUST00000009425.7
ENSMUST00000204267.3 ENSMUST00000204930.3 ENSMUST00000204182.2 |
Rarres2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr1_+_85528392 | 0.64 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr2_+_157209506 | 0.64 |
ENSMUST00000081202.6
|
Manbal
|
mannosidase, beta A, lysosomal-like |
| chr12_+_31315270 | 0.64 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr3_-_88194517 | 0.64 |
ENSMUST00000165196.8
|
Gm38392
|
predicted gene, 38392 |
| chr19_-_5507532 | 0.64 |
ENSMUST00000236881.2
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr19_-_11243530 | 0.63 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_+_171041583 | 0.62 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_-_69704122 | 0.61 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr4_+_86792633 | 0.61 |
ENSMUST00000045224.14
ENSMUST00000084433.5 |
Acer2
|
alkaline ceramidase 2 |
| chr11_-_16958647 | 0.61 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr17_+_28426831 | 0.61 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr2_+_25293140 | 0.60 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr11_+_32176481 | 0.60 |
ENSMUST00000020528.14
|
Mpg
|
N-methylpurine-DNA glycosylase |
| chr4_-_41640321 | 0.60 |
ENSMUST00000127306.2
|
Enho
|
energy homeostasis associated |
| chr7_+_3339059 | 0.60 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr5_+_142946098 | 0.60 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr11_+_59839032 | 0.60 |
ENSMUST00000081980.7
|
Med9
|
mediator complex subunit 9 |
| chr2_+_35172392 | 0.59 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
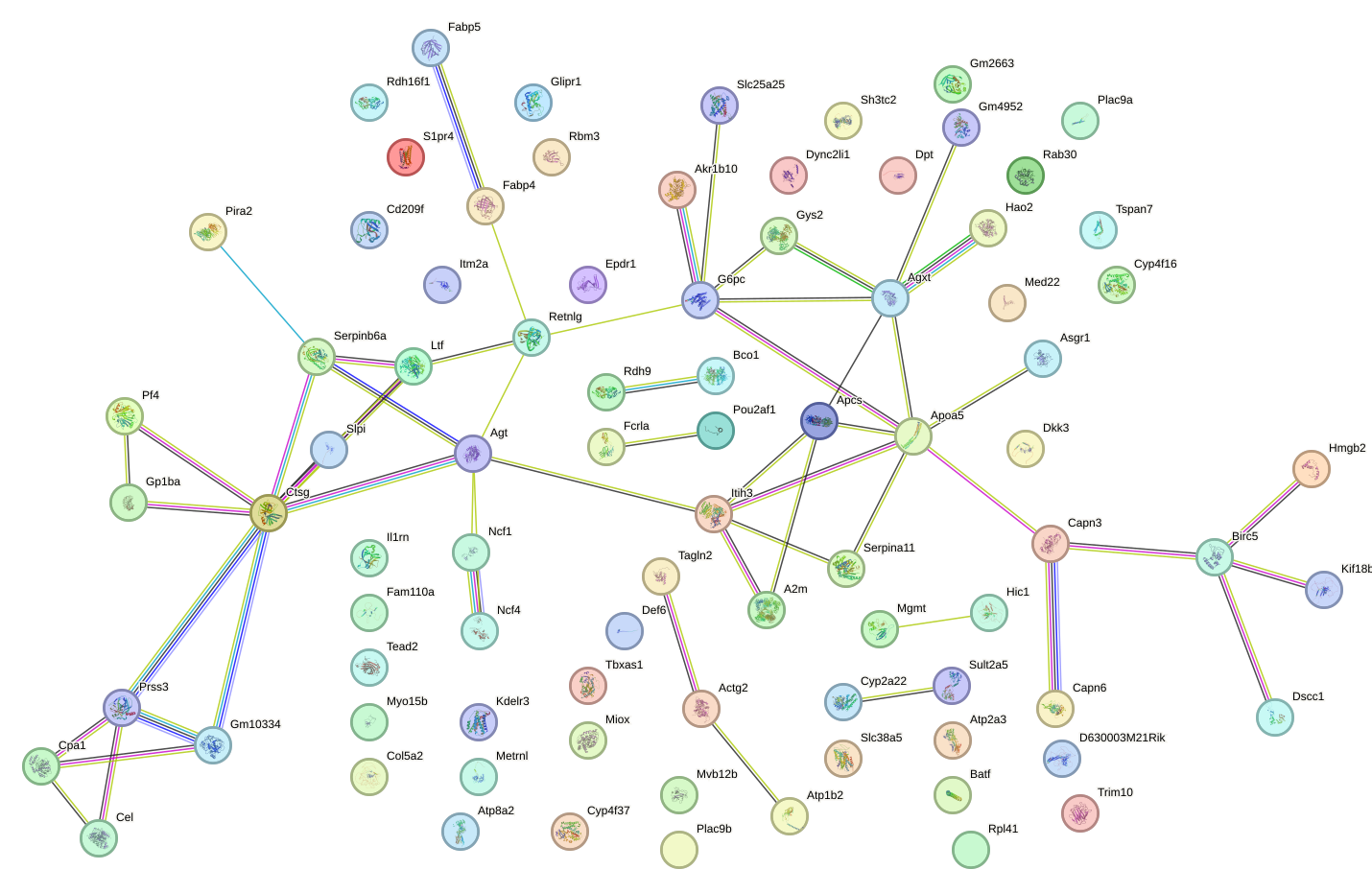
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.0 | 3.0 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.7 | 2.9 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.6 | 2.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.6 | 1.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 | 1.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 2.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.3 | 2.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 0.3 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.3 | 0.9 | GO:0034226 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.3 | 0.9 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 2.1 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.3 | 2.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 2.2 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 | 1.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.9 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.2 | 2.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.9 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.2 | 0.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.9 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.2 | 0.7 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 | 0.6 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 0.6 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.2 | 1.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.8 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.2 | 1.0 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.2 | 0.8 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 0.8 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 0.5 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.2 | 2.9 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 0.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 3.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 0.7 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 0.7 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 1.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 1.5 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.2 | 0.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.5 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.2 | 2.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:1903921 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 1.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 1.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.6 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.7 | GO:0007494 | midgut development(GO:0007494) |
| 0.1 | 0.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 1.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 2.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.3 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.8 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.5 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.4 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.5 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 1.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 2.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 1.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.7 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 1.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.8 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.3 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.1 | 0.5 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 1.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.2 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) chemorepulsion of axon(GO:0061643) |
| 0.1 | 0.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.2 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.1 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 3.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.5 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.4 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 2.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 1.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.5 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 1.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 1.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.9 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 6.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 3.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.0 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 3.3 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.0 | 0.2 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.5 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 1.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 1.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.3 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 4.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 1.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 1.9 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0033034 | positive regulation of neutrophil apoptotic process(GO:0033031) positive regulation of myeloid cell apoptotic process(GO:0033034) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 1.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.9 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.2 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.7 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0002719 | negative regulation of production of molecular mediator of immune response(GO:0002701) negative regulation of cytokine production involved in immune response(GO:0002719) |
| 0.0 | 0.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.5 | GO:0010596 | negative regulation of endothelial cell migration(GO:0010596) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.0 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.9 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 1.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.3 | 1.4 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.3 | 1.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 4.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 0.8 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.6 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 1.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 2.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.1 | GO:1990712 | extrinsic component of external side of plasma membrane(GO:0031232) HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 15.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 5.9 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.7 | 2.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.7 | 4.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 1.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.5 | 3.6 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.5 | 2.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 2.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 1.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.4 | 3.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 3.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 1.0 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.3 | 1.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.3 | 1.0 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.3 | 1.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 0.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 1.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 1.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 2.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 3.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 2.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 1.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 2.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.5 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.2 | 1.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 0.8 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 1.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 3.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 1.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 5.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.6 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 3.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.2 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.7 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 2.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.5 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 10.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 7.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 4.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 6.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 4.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |