Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
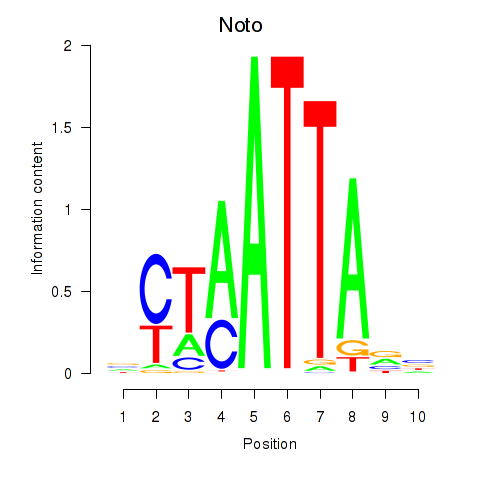
Results for Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 0.41
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
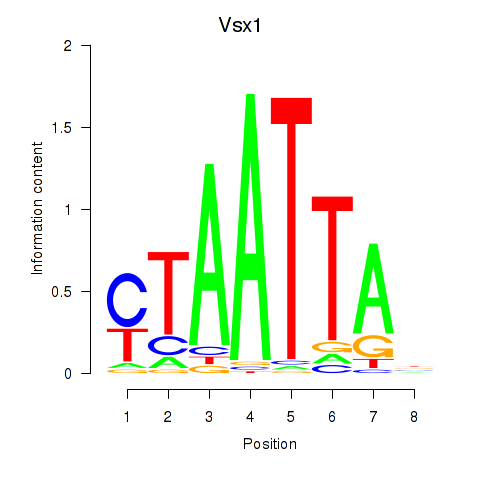
Vsx1
|
ENSMUSG00000033080.10 | visual system homeobox 1 |
|
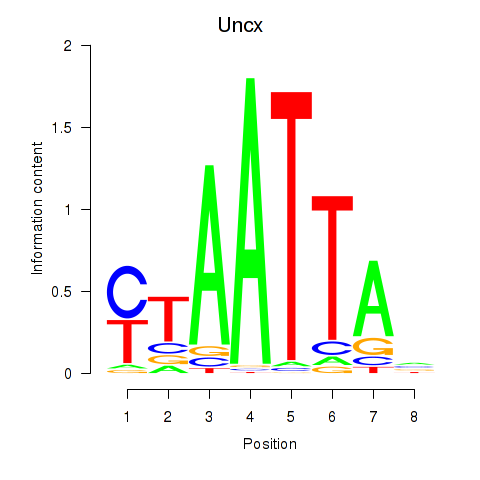
Uncx
|
ENSMUSG00000029546.13 | UNC homeobox |
|
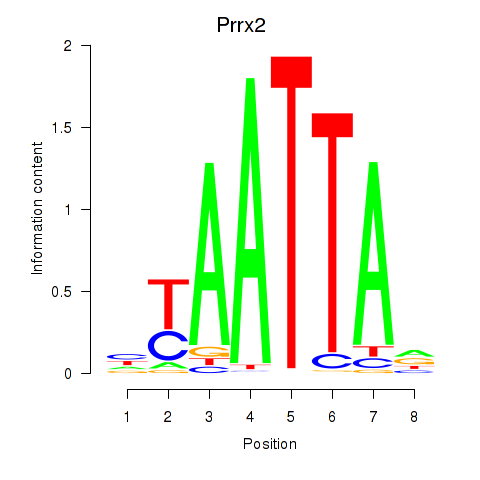
Prrx2
|
ENSMUSG00000039476.14 | paired related homeobox 2 |
|
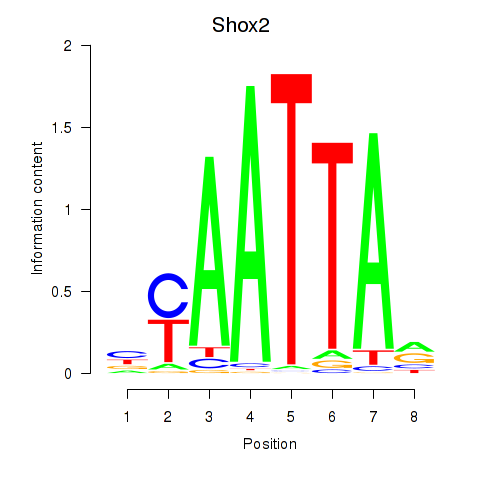
Shox2
|
ENSMUSG00000027833.17 | short stature homeobox 2 |
|
Noto
|
ENSMUSG00000068302.9 | notochord homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Shox2 | mm39_v1_chr3_-_66888474_66888651 | -0.28 | 9.2e-02 | Click! |
| Prrx2 | mm39_v1_chr2_+_30724984_30724984 | -0.10 | 5.7e-01 | Click! |
| Uncx | mm39_v1_chr5_+_139529643_139529662 | -0.07 | 7.0e-01 | Click! |
| Vsx1 | mm39_v1_chr2_-_150531280_150531280 | 0.02 | 9.0e-01 | Click! |
Activity profile of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Sorted Z-values of Vsx1_Uncx_Prrx2_Shox2_Noto motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_138056914 | 2.44 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr14_-_118289557 | 0.84 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr12_-_25147139 | 0.80 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr5_+_115373895 | 0.77 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr5_+_14075281 | 0.75 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chrX_+_132751729 | 0.58 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr4_-_14621805 | 0.53 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr9_+_118307412 | 0.51 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr5_-_77262968 | 0.50 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr4_-_14621669 | 0.50 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_30936013 | 0.48 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr3_+_59989282 | 0.46 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr6_-_41752111 | 0.46 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr14_-_64654397 | 0.44 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chrM_+_2743 | 0.44 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr6_-_115569504 | 0.44 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr9_+_118307250 | 0.43 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr18_+_57601541 | 0.43 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr1_+_74324089 | 0.43 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr8_-_5155347 | 0.41 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr2_+_124978518 | 0.40 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr14_+_26722319 | 0.40 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr4_-_154721288 | 0.38 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr14_-_68771138 | 0.38 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr4_-_97666279 | 0.38 |
ENSMUST00000146447.8
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr6_-_147165623 | 0.38 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chr14_-_64654592 | 0.38 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr4_+_109835224 | 0.37 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr10_+_127734384 | 0.37 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr11_-_87249837 | 0.37 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr4_+_135870808 | 0.36 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chrX_-_142716085 | 0.36 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr15_+_92495007 | 0.36 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr18_+_56565188 | 0.35 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr2_+_124978612 | 0.34 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr7_+_51528788 | 0.34 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr15_+_39522905 | 0.34 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr8_-_3675274 | 0.34 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr7_+_114344920 | 0.33 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr7_-_99512558 | 0.33 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr3_+_93301003 | 0.32 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr2_+_27055245 | 0.32 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr5_+_104350475 | 0.32 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr13_-_103042554 | 0.32 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_85763545 | 0.32 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr7_-_12829100 | 0.32 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chrM_+_7006 | 0.31 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr10_+_128173603 | 0.30 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr11_+_116734104 | 0.30 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chrX_+_110808231 | 0.30 |
ENSMUST00000207546.2
|
Gm45194
|
predicted gene 45194 |
| chr10_+_50770836 | 0.30 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr8_-_3674993 | 0.30 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr4_-_14621497 | 0.29 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chrX_+_149330371 | 0.29 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr11_-_59466995 | 0.29 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr18_+_23548192 | 0.28 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr11_+_70410009 | 0.28 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr7_-_44752508 | 0.28 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr13_-_53627110 | 0.27 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr13_-_43634695 | 0.27 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chrX_+_159551009 | 0.27 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr1_-_158183894 | 0.27 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr13_-_103042294 | 0.26 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_24194729 | 0.26 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr11_+_70410445 | 0.26 |
ENSMUST00000179000.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr10_+_101994841 | 0.26 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr7_+_65343156 | 0.26 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr1_-_163552693 | 0.26 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr14_-_36820304 | 0.26 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr3_+_125197722 | 0.25 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr17_-_84154196 | 0.25 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr5_-_86521273 | 0.25 |
ENSMUST00000031175.12
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr11_-_43792013 | 0.25 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr18_+_24737009 | 0.25 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr2_+_9887427 | 0.25 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr7_-_49286594 | 0.25 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr16_+_43323970 | 0.24 |
ENSMUST00000126100.8
ENSMUST00000123047.8 ENSMUST00000156981.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_+_55448432 | 0.24 |
ENSMUST00000034869.11
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr3_+_108000425 | 0.24 |
ENSMUST00000151326.8
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr14_-_109151590 | 0.24 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr17_-_84154173 | 0.24 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr12_+_111780604 | 0.24 |
ENSMUST00000021714.9
ENSMUST00000223211.2 ENSMUST00000222843.2 ENSMUST00000221375.2 |
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr11_+_6366259 | 0.23 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr5_+_139529643 | 0.23 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr9_-_37580478 | 0.23 |
ENSMUST00000011262.4
|
Panx3
|
pannexin 3 |
| chr13_+_118851214 | 0.23 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr5_+_108842294 | 0.23 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr12_-_11258973 | 0.23 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr16_-_45544960 | 0.22 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr19_-_13827773 | 0.22 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr8_-_3675024 | 0.22 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr15_-_50753792 | 0.22 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_145811028 | 0.22 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr1_-_74924481 | 0.21 |
ENSMUST00000159232.2
ENSMUST00000068631.4 |
Fev
|
FEV transcription factor, ETS family member |
| chr13_+_22563988 | 0.21 |
ENSMUST00000227685.2
ENSMUST00000227689.2 ENSMUST00000227846.2 |
Vmn1r199
|
vomeronasal 1 receptor 199 |
| chr11_+_115225557 | 0.21 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chrX_+_159551171 | 0.21 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr5_+_35156454 | 0.21 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr19_-_13828056 | 0.21 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr18_-_81029986 | 0.21 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr15_-_103123711 | 0.20 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr15_-_34356567 | 0.20 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr3_+_55369149 | 0.20 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr10_-_12689345 | 0.20 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr1_-_14374842 | 0.20 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chrX_-_142716200 | 0.20 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr13_-_23041731 | 0.20 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr12_+_38830812 | 0.20 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr1_-_72323464 | 0.19 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr15_+_21111428 | 0.19 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr17_+_17622934 | 0.19 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr1_-_14374794 | 0.19 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_9369463 | 0.19 |
ENSMUST00000140295.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr18_-_3280999 | 0.19 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr1_-_9368721 | 0.19 |
ENSMUST00000132064.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr2_-_17465410 | 0.18 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr18_+_23548534 | 0.18 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr8_-_32408864 | 0.18 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr1_-_9369045 | 0.18 |
ENSMUST00000191683.6
|
Sntg1
|
syntrophin, gamma 1 |
| chr8_-_62576140 | 0.18 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr3_+_68479578 | 0.18 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_+_43430459 | 0.18 |
ENSMUST00000014058.11
|
Klk10
|
kallikrein related-peptidase 10 |
| chr7_+_49559859 | 0.18 |
ENSMUST00000056442.12
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr2_+_162829422 | 0.17 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_72875187 | 0.17 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr7_+_138828391 | 0.17 |
ENSMUST00000093993.5
ENSMUST00000172136.9 |
Pwwp2b
|
PWWP domain containing 2B |
| chr2_+_83554770 | 0.17 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr3_-_154760978 | 0.17 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr16_+_42727926 | 0.17 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr18_+_23548455 | 0.17 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr1_+_88234454 | 0.17 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr7_-_115423934 | 0.17 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_-_58499398 | 0.17 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr2_+_162829250 | 0.17 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_-_46316767 | 0.17 |
ENSMUST00000168335.3
ENSMUST00000107669.9 |
Tph1
|
tryptophan hydroxylase 1 |
| chr4_+_97665992 | 0.17 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr13_+_8252895 | 0.17 |
ENSMUST00000064473.13
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr4_-_155141241 | 0.17 |
ENSMUST00000131173.3
|
Plch2
|
phospholipase C, eta 2 |
| chr3_-_92031247 | 0.16 |
ENSMUST00000070284.4
|
Prr9
|
proline rich 9 |
| chr7_+_99512701 | 0.16 |
ENSMUST00000207855.2
|
Xrra1
|
X-ray radiation resistance associated 1 |
| chr2_-_63014514 | 0.16 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr10_+_73782857 | 0.16 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr14_-_110992533 | 0.16 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr2_-_18053595 | 0.16 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr2_-_87543523 | 0.16 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chr6_+_145879839 | 0.16 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr19_-_42117420 | 0.16 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chr6_+_8948608 | 0.16 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr5_-_66672158 | 0.16 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_-_72323407 | 0.16 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_+_104696235 | 0.16 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr10_-_107330580 | 0.16 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr4_-_110149916 | 0.15 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr7_-_37472290 | 0.15 |
ENSMUST00000176205.8
|
Zfp536
|
zinc finger protein 536 |
| chr13_+_8252957 | 0.15 |
ENSMUST00000123187.2
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr19_-_11816583 | 0.15 |
ENSMUST00000214887.2
|
Olfr1417
|
olfactory receptor 1417 |
| chr4_+_13751297 | 0.15 |
ENSMUST00000105566.9
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr13_-_21798192 | 0.15 |
ENSMUST00000051874.6
|
Olfr1362
|
olfactory receptor 1362 |
| chr13_+_22508759 | 0.15 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr13_+_111391544 | 0.15 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr17_+_49922129 | 0.15 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr9_+_96140750 | 0.15 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr6_+_63232955 | 0.15 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr1_+_53100796 | 0.15 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr11_-_99412162 | 0.15 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr8_+_23114035 | 0.15 |
ENSMUST00000033936.8
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_-_14380327 | 0.14 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr6_+_29853745 | 0.14 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr11_-_99412084 | 0.14 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr10_+_126911134 | 0.14 |
ENSMUST00000239120.2
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_+_108145802 | 0.14 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr2_+_177969456 | 0.14 |
ENSMUST00000133267.3
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr15_-_101482320 | 0.14 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr3_-_86455575 | 0.14 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr6_-_138404076 | 0.14 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr19_+_59446804 | 0.14 |
ENSMUST00000062216.4
|
Emx2
|
empty spiracles homeobox 2 |
| chr3_-_50398027 | 0.14 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chrX_-_156198282 | 0.14 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr8_-_32440071 | 0.14 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr2_+_61634797 | 0.14 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr10_+_101994719 | 0.14 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr8_+_46111703 | 0.14 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_92446141 | 0.14 |
ENSMUST00000189174.2
|
Olfr1414
|
olfactory receptor 1414 |
| chr2_+_57887896 | 0.14 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr8_-_85573489 | 0.14 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr2_-_86857424 | 0.14 |
ENSMUST00000214857.2
ENSMUST00000215972.2 |
Olfr1104
|
olfactory receptor 1104 |
| chr11_+_94218810 | 0.13 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr13_+_23431304 | 0.13 |
ENSMUST00000235331.2
ENSMUST00000236495.2 ENSMUST00000238002.2 |
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr5_+_104318542 | 0.13 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr12_+_38831093 | 0.13 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr8_+_121842902 | 0.13 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr15_-_66985760 | 0.13 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr18_-_81029751 | 0.13 |
ENSMUST00000238808.2
|
Sall3
|
spalt like transcription factor 3 |
| chr7_-_37472979 | 0.13 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr3_-_130524024 | 0.13 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr2_-_86109346 | 0.13 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr10_-_23977810 | 0.13 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr1_+_161322219 | 0.13 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr18_-_57108405 | 0.13 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr1_+_172383499 | 0.13 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx1_Uncx_Prrx2_Shox2_Noto
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 0.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 1.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.4 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.2 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.2 | GO:0048880 | hindbrain tangential cell migration(GO:0021934) sensory system development(GO:0048880) |
| 0.1 | 0.3 | GO:2000293 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.5 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.4 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:2000722 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 0.1 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0072144 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0090336 | negative regulation of granulocyte differentiation(GO:0030853) positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.1 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.0 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 0.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.3 | GO:0016232 | HNK-1 sulfotransferase activity(GO:0016232) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.2 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 1.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.0 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |