Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
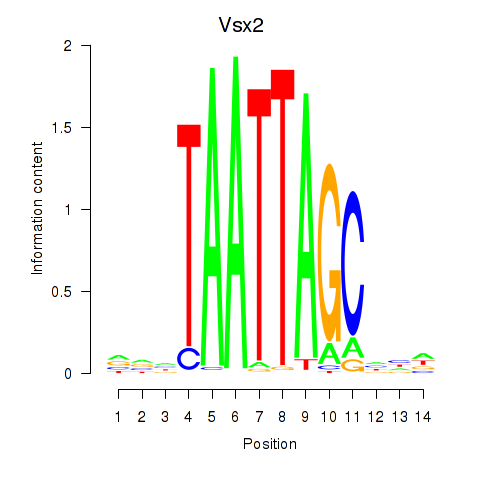
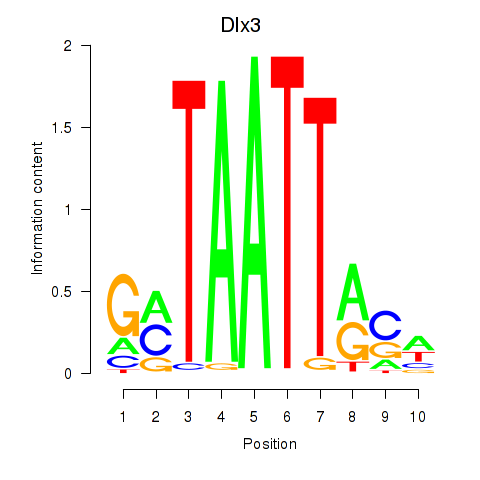
Results for Vsx2_Dlx3
Z-value: 0.40
Transcription factors associated with Vsx2_Dlx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vsx2
|
ENSMUSG00000021239.13 | visual system homeobox 2 |
|
Dlx3
|
ENSMUSG00000001510.9 | distal-less homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vsx2 | mm39_v1_chr12_+_84616536_84616614 | -0.24 | 1.6e-01 | Click! |
| Dlx3 | mm39_v1_chr11_+_95010935_95010954 | 0.19 | 2.7e-01 | Click! |
Activity profile of Vsx2_Dlx3 motif
Sorted Z-values of Vsx2_Dlx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_130524024 | 1.36 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr6_+_30541581 | 1.25 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr2_+_84564394 | 1.08 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr3_-_130523954 | 1.05 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr2_-_168609110 | 0.95 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr18_-_43610829 | 0.76 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr2_-_168608949 | 0.73 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr3_+_51568588 | 0.70 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr14_+_26722319 | 0.69 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_+_139857640 | 0.52 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr8_-_3675274 | 0.51 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chrX_+_139857688 | 0.51 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr7_-_115630282 | 0.49 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr3_-_116047148 | 0.47 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr2_-_153079828 | 0.42 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr7_-_128020397 | 0.41 |
ENSMUST00000147840.2
|
Rgs10
|
regulator of G-protein signalling 10 |
| chr7_-_100504610 | 0.41 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr19_-_46033353 | 0.39 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr12_+_108572015 | 0.33 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr8_-_3675525 | 0.27 |
ENSMUST00000144977.2
ENSMUST00000136105.8 ENSMUST00000128566.8 |
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr10_+_128173603 | 0.26 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr8_-_3674993 | 0.25 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr17_-_71305003 | 0.25 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_-_111843053 | 0.24 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrX_+_111003193 | 0.24 |
ENSMUST00000130247.9
ENSMUST00000038546.7 |
Tex16
|
testis expressed gene 16 |
| chr7_-_115445352 | 0.24 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr8_-_3675024 | 0.24 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_+_127750978 | 0.23 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr1_+_172383499 | 0.22 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr14_+_76741625 | 0.22 |
ENSMUST00000177207.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr3_-_88317601 | 0.21 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr14_+_99536111 | 0.20 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr18_+_57601541 | 0.20 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr12_-_114398864 | 0.20 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr12_+_52746158 | 0.19 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr6_-_30936013 | 0.19 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr6_-_51443602 | 0.19 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr15_+_91722524 | 0.18 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr6_-_102441628 | 0.17 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr5_-_65855511 | 0.17 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr7_-_115445315 | 0.17 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr18_+_82932747 | 0.17 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr1_-_79417732 | 0.17 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chrX_+_132751729 | 0.16 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr15_-_101602734 | 0.16 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr5_-_123127346 | 0.15 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_+_116734104 | 0.15 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr3_-_151953894 | 0.14 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chrX_+_158623460 | 0.14 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr1_+_53100796 | 0.14 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr12_-_11258973 | 0.14 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr12_-_104439589 | 0.14 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr4_-_154721288 | 0.14 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr14_+_76741918 | 0.14 |
ENSMUST00000022587.10
ENSMUST00000134109.2 |
Tsc22d1
|
TSC22 domain family, member 1 |
| chr10_-_23977810 | 0.13 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr2_-_111880531 | 0.13 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr12_-_114321838 | 0.13 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr14_+_32043944 | 0.13 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr7_-_115459082 | 0.13 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_14075281 | 0.13 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr13_+_118851214 | 0.13 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr6_+_106095726 | 0.12 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr13_-_74882374 | 0.12 |
ENSMUST00000220738.2
|
Cast
|
calpastatin |
| chr5_-_84565218 | 0.12 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr4_-_132237804 | 0.12 |
ENSMUST00000030724.9
|
Sesn2
|
sestrin 2 |
| chr10_-_128918779 | 0.12 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr14_+_50360643 | 0.11 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr5_-_137530214 | 0.11 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr4_+_133795740 | 0.11 |
ENSMUST00000121391.8
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr9_+_53678801 | 0.11 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr2_-_79287095 | 0.11 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr3_+_159545309 | 0.11 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr4_-_41464816 | 0.11 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr15_+_44291470 | 0.10 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr2_-_86109346 | 0.10 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr17_-_21006419 | 0.10 |
ENSMUST00000233605.2
ENSMUST00000232812.2 ENSMUST00000233186.2 ENSMUST00000233164.2 |
Vmn1r228
|
vomeronasal 1 receptor 228 |
| chr4_+_5724305 | 0.10 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr3_-_144514386 | 0.10 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr1_-_92446383 | 0.10 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr11_+_101877876 | 0.10 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr18_+_23548534 | 0.10 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr13_-_74882328 | 0.10 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr5_+_4073343 | 0.10 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr12_-_111780268 | 0.10 |
ENSMUST00000021715.6
|
Xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr16_-_34083549 | 0.09 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr1_-_171854818 | 0.09 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr5_+_107656810 | 0.09 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr4_+_110254907 | 0.09 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr4_-_110143777 | 0.09 |
ENSMUST00000138972.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr3_-_50398027 | 0.09 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr1_-_173018204 | 0.09 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr2_+_36120438 | 0.09 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_-_47680528 | 0.09 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr12_+_99930757 | 0.08 |
ENSMUST00000160413.8
ENSMUST00000177549.8 ENSMUST00000049788.9 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr1_-_92446141 | 0.08 |
ENSMUST00000189174.2
|
Olfr1414
|
olfactory receptor 1414 |
| chr4_-_82970467 | 0.08 |
ENSMUST00000107230.8
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr16_-_89615225 | 0.08 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr2_-_7400690 | 0.08 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_+_110254858 | 0.08 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr15_+_6673167 | 0.08 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr18_-_81029986 | 0.08 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr2_+_89708781 | 0.08 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr17_+_35844091 | 0.08 |
ENSMUST00000025273.9
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr18_+_23548192 | 0.08 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr5_-_137529465 | 0.08 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr2_-_168607166 | 0.08 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr9_+_19716202 | 0.08 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr9_-_96513529 | 0.07 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr4_-_82970384 | 0.07 |
ENSMUST00000170248.9
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr1_-_134883645 | 0.07 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_38830812 | 0.07 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr7_-_102566717 | 0.07 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr4_+_109835224 | 0.07 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr9_+_20209828 | 0.07 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr5_-_116427003 | 0.07 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr7_-_103094646 | 0.07 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr17_-_21216726 | 0.07 |
ENSMUST00000237195.2
ENSMUST00000237629.2 ENSMUST00000056339.3 |
Vmn1r233
|
vomeronasal 1 receptor 233 |
| chr17_+_17622934 | 0.07 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr11_+_102175985 | 0.07 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr12_-_56581823 | 0.07 |
ENSMUST00000178477.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr4_+_43851565 | 0.06 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr5_-_70999547 | 0.06 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chrX_-_48980360 | 0.06 |
ENSMUST00000217355.3
|
Olfr1322
|
olfactory receptor 1322 |
| chr9_-_106768601 | 0.06 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr5_+_115373895 | 0.06 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr9_-_119897358 | 0.06 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr15_-_34356567 | 0.06 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr18_+_23548455 | 0.06 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr17_-_81372483 | 0.06 |
ENSMUST00000025093.6
|
Thumpd2
|
THUMP domain containing 2 |
| chr1_-_134883577 | 0.06 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_38831093 | 0.06 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr16_-_92196954 | 0.06 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr10_-_28862289 | 0.06 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chrX_+_133088238 | 0.06 |
ENSMUST00000064476.5
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr5_+_27466914 | 0.06 |
ENSMUST00000101471.4
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr5_-_116162415 | 0.05 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr4_-_110149916 | 0.05 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr6_+_97968737 | 0.05 |
ENSMUST00000043628.13
ENSMUST00000203938.2 |
Mitf
|
melanogenesis associated transcription factor |
| chr15_+_21111428 | 0.05 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr2_+_69050315 | 0.05 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr5_-_138185438 | 0.05 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr5_-_138185686 | 0.05 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr14_+_54183465 | 0.05 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr4_-_52919172 | 0.05 |
ENSMUST00000107667.3
ENSMUST00000213989.2 |
Olfr272
|
olfactory receptor 272 |
| chr19_-_13828056 | 0.05 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chrX_+_105626790 | 0.05 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr14_-_52277310 | 0.05 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chr19_-_5610628 | 0.04 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr13_-_112698563 | 0.04 |
ENSMUST00000224510.2
|
Il31ra
|
interleukin 31 receptor A |
| chr6_+_63232955 | 0.04 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr8_+_10299288 | 0.04 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr19_-_13827773 | 0.04 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr7_-_102507962 | 0.04 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr10_+_69623262 | 0.04 |
ENSMUST00000183240.2
|
Ank3
|
ankyrin 3, epithelial |
| chr16_+_34511073 | 0.04 |
ENSMUST00000231609.2
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr1_+_173983199 | 0.04 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr2_+_9887427 | 0.04 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr17_-_37472385 | 0.04 |
ENSMUST00000219235.3
|
Olfr93
|
olfactory receptor 93 |
| chr9_-_119897328 | 0.04 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr2_+_49677688 | 0.04 |
ENSMUST00000028103.13
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr6_+_134958681 | 0.04 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chrX_-_142716085 | 0.04 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr6_-_136758716 | 0.04 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr5_-_143279378 | 0.04 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr5_+_139529643 | 0.04 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr17_+_93506590 | 0.04 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr17_+_93506435 | 0.04 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr8_+_57774010 | 0.03 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_+_8690398 | 0.03 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr16_+_27126239 | 0.03 |
ENSMUST00000066852.9
|
Ostn
|
osteocrin |
| chr2_+_127696548 | 0.03 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr3_+_137770813 | 0.03 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr2_-_7400780 | 0.03 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_65160767 | 0.03 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr19_+_38121214 | 0.03 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr17_-_36008863 | 0.03 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr12_-_55061117 | 0.03 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr18_+_69479211 | 0.03 |
ENSMUST00000201235.4
|
Tcf4
|
transcription factor 4 |
| chr1_-_158183894 | 0.03 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr8_-_32440071 | 0.03 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr17_+_38110779 | 0.03 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr13_-_103042554 | 0.03 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_99412162 | 0.03 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr1_-_179527845 | 0.03 |
ENSMUST00000223392.2
|
Kif28
|
kinesin family member 28 |
| chr11_-_102076028 | 0.03 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr11_+_115225557 | 0.03 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr9_-_21223551 | 0.03 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr7_+_103199575 | 0.02 |
ENSMUST00000106888.3
|
Olfr613
|
olfactory receptor 613 |
| chrX_-_142716200 | 0.02 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_+_88644840 | 0.02 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr11_-_102469896 | 0.02 |
ENSMUST00000107080.2
|
Gm11627
|
predicted gene 11627 |
| chr2_-_7400997 | 0.02 |
ENSMUST00000137733.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr10_-_6930376 | 0.02 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_-_21798192 | 0.02 |
ENSMUST00000051874.6
|
Olfr1362
|
olfactory receptor 1362 |
| chr2_+_153742294 | 0.02 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr6_-_68968278 | 0.02 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr1_-_92412835 | 0.02 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr4_-_45532470 | 0.02 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr12_-_56660054 | 0.02 |
ENSMUST00000072631.6
|
Nkx2-9
|
NK2 homeobox 9 |
| chr16_+_43574389 | 0.02 |
ENSMUST00000229953.2
|
Drd3
|
dopamine receptor D3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx2_Dlx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 1.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 1.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 1.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.7 | GO:0019370 | glutathione biosynthetic process(GO:0006750) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.2 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.8 | GO:0000792 | heterochromatin(GO:0000792) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |