Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Xbp1_Creb3l1
Z-value: 1.20
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
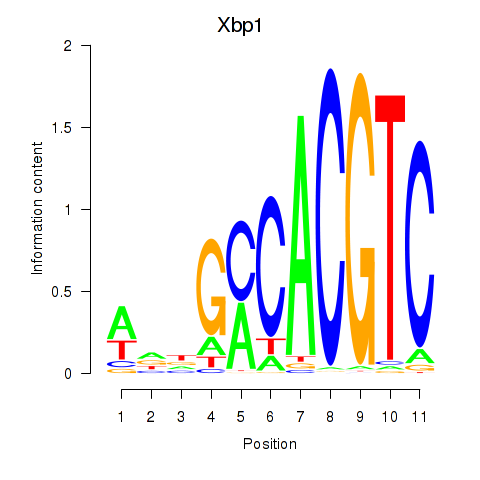
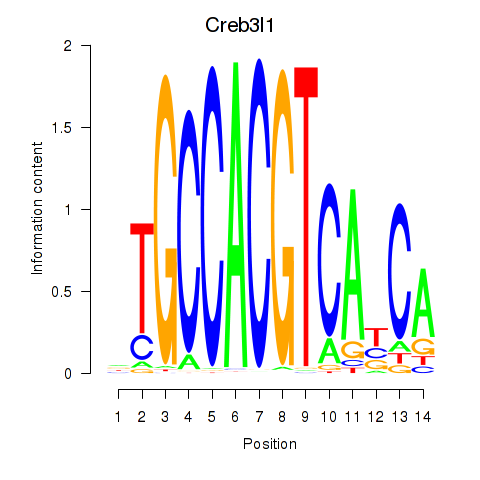
Xbp1
|
ENSMUSG00000020484.20 | X-box binding protein 1 |
|
Creb3l1
|
ENSMUSG00000027230.10 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | mm39_v1_chr2_-_91854844_91854847 | 0.48 | 2.9e-03 | Click! |
| Xbp1 | mm39_v1_chr11_+_5470968_5470996 | 0.47 | 3.9e-03 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44741622 | 3.88 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_44741609 | 3.55 |
ENSMUST00000210734.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_+_128744689 | 2.61 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr10_+_128745214 | 2.58 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr1_-_93406091 | 2.49 |
ENSMUST00000188165.2
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr9_+_103940575 | 2.42 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr2_-_168584020 | 2.39 |
ENSMUST00000109177.8
|
Atp9a
|
ATPase, class II, type 9A |
| chr9_-_86453862 | 2.31 |
ENSMUST00000070064.11
ENSMUST00000072585.8 |
Pgm3
|
phosphoglucomutase 3 |
| chr15_+_79400597 | 2.27 |
ENSMUST00000010974.9
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr19_+_32573182 | 2.24 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr1_+_157334347 | 2.22 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr4_-_118401185 | 2.22 |
ENSMUST00000128098.8
|
Tmem125
|
transmembrane protein 125 |
| chr1_+_157334298 | 2.20 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr13_-_38712387 | 2.19 |
ENSMUST00000035988.16
|
Txndc5
|
thioredoxin domain containing 5 |
| chr1_-_93406515 | 2.06 |
ENSMUST00000170883.8
ENSMUST00000189025.7 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr3_-_27764571 | 2.04 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr14_+_20724378 | 1.84 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr7_-_44741138 | 1.79 |
ENSMUST00000210527.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr19_-_10079091 | 1.78 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr2_+_80145805 | 1.76 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr3_-_97675231 | 1.74 |
ENSMUST00000045243.15
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr14_-_60324265 | 1.72 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr18_+_57487908 | 1.64 |
ENSMUST00000238069.2
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr10_-_128236317 | 1.62 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr4_-_155737841 | 1.61 |
ENSMUST00000030937.2
|
Mmp23
|
matrix metallopeptidase 23 |
| chr11_-_96807192 | 1.53 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr6_+_29348068 | 1.40 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr7_-_34914675 | 1.37 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr6_+_4505493 | 1.35 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr14_+_20724366 | 1.33 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr19_+_3758285 | 1.27 |
ENSMUST00000237320.2
ENSMUST00000039048.2 ENSMUST00000235295.2 ENSMUST00000237955.2 ENSMUST00000235837.2 |
1810055G02Rik
|
RIKEN cDNA 1810055G02 gene |
| chr10_+_41395870 | 1.26 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr4_-_108263873 | 1.21 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr1_-_10038030 | 1.20 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr3_+_123061094 | 1.19 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr18_+_57487800 | 1.19 |
ENSMUST00000025490.10
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr11_-_96807233 | 1.18 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_+_78079631 | 1.18 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr10_-_128047658 | 1.18 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chr8_+_26210064 | 1.17 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr3_-_27764522 | 1.16 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_+_150939521 | 1.12 |
ENSMUST00000030811.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr14_+_26359191 | 1.08 |
ENSMUST00000022429.9
|
Arf4
|
ADP-ribosylation factor 4 |
| chr2_+_150628655 | 1.08 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr9_-_44679136 | 1.04 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr4_-_57300361 | 1.04 |
ENSMUST00000153926.8
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr15_+_31224616 | 1.03 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr6_-_88495835 | 0.98 |
ENSMUST00000032168.7
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr10_+_42637479 | 0.97 |
ENSMUST00000019937.5
|
Sec63
|
SEC63-like (S. cerevisiae) |
| chr17_-_34250474 | 0.96 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr10_-_128236366 | 0.95 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr2_-_143853122 | 0.93 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr17_-_34250616 | 0.93 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr5_+_113873864 | 0.92 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chrX_-_135642025 | 0.89 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr5_+_31026967 | 0.89 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr14_+_26359390 | 0.88 |
ENSMUST00000112318.10
|
Arf4
|
ADP-ribosylation factor 4 |
| chr1_+_171910073 | 0.87 |
ENSMUST00000135192.8
|
Copa
|
coatomer protein complex subunit alpha |
| chr19_+_5138562 | 0.87 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr11_+_97690585 | 0.86 |
ENSMUST00000129558.8
|
Lasp1
|
LIM and SH3 protein 1 |
| chr2_+_70491901 | 0.86 |
ENSMUST00000112201.8
ENSMUST00000028509.11 ENSMUST00000133432.8 ENSMUST00000112205.2 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr8_-_106863423 | 0.86 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr4_+_43562706 | 0.85 |
ENSMUST00000167751.2
ENSMUST00000132631.2 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr1_+_171910343 | 0.80 |
ENSMUST00000027833.12
|
Copa
|
coatomer protein complex subunit alpha |
| chr5_+_65127412 | 0.80 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr1_-_171910324 | 0.77 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr19_+_11943265 | 0.77 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein |
| chr11_-_96807273 | 0.76 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_+_63673843 | 0.73 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr1_-_183150867 | 0.70 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr4_-_134431534 | 0.69 |
ENSMUST00000054096.13
ENSMUST00000038628.4 |
Man1c1
|
mannosidase, alpha, class 1C, member 1 |
| chr7_-_121666486 | 0.68 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr16_-_56706494 | 0.68 |
ENSMUST00000023435.6
|
Tmem45a
|
transmembrane protein 45a |
| chr2_+_15060051 | 0.68 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr10_-_24587884 | 0.68 |
ENSMUST00000135846.2
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr4_-_47474283 | 0.67 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr19_+_8875459 | 0.66 |
ENSMUST00000096246.5
ENSMUST00000235274.2 |
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chr15_-_57755753 | 0.65 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr4_+_43441939 | 0.65 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr17_+_34341766 | 0.63 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr10_+_41395410 | 0.63 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr16_-_32698091 | 0.63 |
ENSMUST00000119810.2
|
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr15_+_31224555 | 0.59 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr6_-_54570124 | 0.59 |
ENSMUST00000046520.13
|
Fkbp14
|
FK506 binding protein 14 |
| chr15_+_31224460 | 0.58 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr4_+_137321451 | 0.58 |
ENSMUST00000105840.8
ENSMUST00000105839.8 ENSMUST00000055131.13 ENSMUST00000105838.8 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr17_-_26004298 | 0.56 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr4_-_137523659 | 0.56 |
ENSMUST00000030551.11
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chrX_-_135641869 | 0.56 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr3_-_65299967 | 0.55 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr5_-_35836761 | 0.55 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr2_-_18397547 | 0.55 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr7_-_118091135 | 0.55 |
ENSMUST00000178344.3
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr11_+_97690391 | 0.54 |
ENSMUST00000043843.12
|
Lasp1
|
LIM and SH3 protein 1 |
| chr1_-_55265925 | 0.53 |
ENSMUST00000027121.15
ENSMUST00000114428.3 |
Rftn2
|
raftlin family member 2 |
| chr7_-_126992776 | 0.52 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr12_+_13319110 | 0.52 |
ENSMUST00000042953.10
|
Nbas
|
neuroblastoma amplified sequence |
| chr8_-_106863521 | 0.51 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr10_-_24588030 | 0.51 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr2_-_155534235 | 0.51 |
ENSMUST00000103140.5
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr8_+_70755168 | 0.51 |
ENSMUST00000066469.14
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr7_-_113853894 | 0.50 |
ENSMUST00000033012.9
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr4_-_129534752 | 0.49 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr3_-_131196213 | 0.48 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr11_-_20781009 | 0.47 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr10_-_84369831 | 0.47 |
ENSMUST00000167671.2
ENSMUST00000053871.5 |
Ckap4
|
cytoskeleton-associated protein 4 |
| chr6_-_52181393 | 0.45 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr16_+_36695479 | 0.45 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr10_+_90918802 | 0.44 |
ENSMUST00000020150.10
ENSMUST00000020149.6 |
Ikbip
|
IKBKB interacting protein |
| chr1_+_119453849 | 0.44 |
ENSMUST00000183952.2
|
Tmem185b
|
transmembrane protein 185B |
| chr7_+_128125339 | 0.44 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr9_-_35030479 | 0.43 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr2_-_105229653 | 0.43 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr11_-_108234743 | 0.42 |
ENSMUST00000059595.11
|
Prkca
|
protein kinase C, alpha |
| chr2_+_168072519 | 0.41 |
ENSMUST00000099071.5
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr10_-_122912272 | 0.40 |
ENSMUST00000219203.2
ENSMUST00000073792.11 |
Mon2
|
MON2 homolog, regulator of endosome to Golgi trafficking |
| chr16_-_35311243 | 0.38 |
ENSMUST00000023550.9
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr9_-_103940247 | 0.37 |
ENSMUST00000035166.12
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr10_+_61516078 | 0.37 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr16_+_32698149 | 0.36 |
ENSMUST00000023489.11
ENSMUST00000171325.9 |
Fyttd1
|
forty-two-three domain containing 1 |
| chr11_-_100861713 | 0.35 |
ENSMUST00000060792.6
|
Cavin1
|
caveolae associated 1 |
| chr17_-_64638887 | 0.33 |
ENSMUST00000172818.8
|
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr7_-_44519196 | 0.33 |
ENSMUST00000046575.17
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr4_-_129534853 | 0.32 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr17_-_64638814 | 0.32 |
ENSMUST00000172733.2
|
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr9_+_57468217 | 0.32 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr11_+_94520567 | 0.31 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr5_+_110324734 | 0.31 |
ENSMUST00000139611.8
ENSMUST00000031477.9 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr9_-_103079312 | 0.31 |
ENSMUST00000035157.10
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr14_-_75991903 | 0.30 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr19_+_46750016 | 0.30 |
ENSMUST00000099373.12
ENSMUST00000077666.6 |
Cnnm2
|
cyclin M2 |
| chr3_-_108469740 | 0.29 |
ENSMUST00000090546.6
|
Tmem167b
|
transmembrane protein 167B |
| chr2_+_105054657 | 0.29 |
ENSMUST00000068813.3
|
Them7
|
thioesterase superfamily member 7 |
| chr6_+_87019819 | 0.29 |
ENSMUST00000113657.8
ENSMUST00000113658.8 ENSMUST00000113655.8 ENSMUST00000032057.8 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr3_-_84489923 | 0.28 |
ENSMUST00000143514.3
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr17_+_34251041 | 0.27 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr17_+_34250757 | 0.27 |
ENSMUST00000044858.16
ENSMUST00000174299.9 |
Rxrb
|
retinoid X receptor beta |
| chr2_-_79259235 | 0.27 |
ENSMUST00000143974.2
|
Cerkl
|
ceramide kinase-like |
| chr5_+_92285748 | 0.27 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr5_+_77122530 | 0.26 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr5_-_72716942 | 0.26 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr11_-_68743845 | 0.25 |
ENSMUST00000108672.2
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr13_+_77856801 | 0.25 |
ENSMUST00000163257.9
ENSMUST00000091459.12 ENSMUST00000099358.5 ENSMUST00000225623.2 ENSMUST00000224908.2 |
Fam172a
|
family with sequence similarity 172, member A |
| chr6_-_72935382 | 0.25 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr4_-_129534403 | 0.24 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr19_-_5821373 | 0.24 |
ENSMUST00000236297.2
ENSMUST00000236773.2 ENSMUST00000025890.10 |
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr18_+_51250748 | 0.24 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr12_+_51424343 | 0.24 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr16_+_51851917 | 0.23 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr6_-_113717689 | 0.23 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr4_+_59003121 | 0.23 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr6_-_72935468 | 0.23 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chrX_+_72830607 | 0.22 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr12_+_110567873 | 0.22 |
ENSMUST00000018851.14
|
Dync1h1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr13_-_76246689 | 0.21 |
ENSMUST00000239063.2
ENSMUST00000120573.3 |
Arsk
|
arylsulfatase K |
| chr16_+_51851948 | 0.21 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_+_126721878 | 0.21 |
ENSMUST00000046765.10
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr6_-_134874778 | 0.20 |
ENSMUST00000165392.8
ENSMUST00000204880.3 ENSMUST00000203409.3 ENSMUST00000046255.14 ENSMUST00000111932.8 ENSMUST00000149375.8 ENSMUST00000116515.9 |
Gpr19
|
G protein-coupled receptor 19 |
| chr3_-_108469468 | 0.20 |
ENSMUST00000106622.3
|
Tmem167b
|
transmembrane protein 167B |
| chr11_+_3464861 | 0.19 |
ENSMUST00000094469.6
|
Selenom
|
selenoprotein M |
| chr10_+_84938452 | 0.19 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr19_-_5821442 | 0.18 |
ENSMUST00000236978.2
|
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr9_+_86454018 | 0.18 |
ENSMUST00000185566.7
ENSMUST00000034988.10 ENSMUST00000179212.3 |
Rwdd2a
|
RWD domain containing 2A |
| chr2_-_38955518 | 0.18 |
ENSMUST00000039165.15
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr7_+_116103599 | 0.18 |
ENSMUST00000032895.15
|
Nucb2
|
nucleobindin 2 |
| chr8_+_71725332 | 0.18 |
ENSMUST00000212935.2
ENSMUST00000170242.8 ENSMUST00000071935.7 ENSMUST00000168839.9 |
Myo9b
|
myosin IXb |
| chr3_-_65300000 | 0.16 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr3_+_96604390 | 0.16 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr4_-_43562397 | 0.16 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr2_+_164339438 | 0.16 |
ENSMUST00000103101.11
ENSMUST00000117066.8 |
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr19_-_6134703 | 0.14 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr6_-_118456198 | 0.14 |
ENSMUST00000161170.2
|
Zfp9
|
zinc finger protein 9 |
| chr1_-_163141278 | 0.13 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr4_-_43000450 | 0.13 |
ENSMUST00000030164.8
|
Vcp
|
valosin containing protein |
| chr5_+_143389573 | 0.13 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr7_+_116103643 | 0.13 |
ENSMUST00000183175.8
|
Nucb2
|
nucleobindin 2 |
| chr2_-_38955452 | 0.12 |
ENSMUST00000112850.9
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr16_+_51851588 | 0.12 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_+_126722113 | 0.12 |
ENSMUST00000212831.2
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chrX_+_72830668 | 0.12 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr19_+_8718837 | 0.11 |
ENSMUST00000177373.8
ENSMUST00000010254.16 |
Stx5a
|
syntaxin 5A |
| chr19_+_8718777 | 0.10 |
ENSMUST00000176381.8
|
Stx5a
|
syntaxin 5A |
| chr7_-_28931873 | 0.10 |
ENSMUST00000085818.6
|
Kcnk6
|
potassium inwardly-rectifying channel, subfamily K, member 6 |
| chr13_+_76246853 | 0.10 |
ENSMUST00000091466.4
ENSMUST00000224386.2 |
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr7_+_26958150 | 0.10 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr6_+_28480336 | 0.09 |
ENSMUST00000001460.14
ENSMUST00000167201.2 |
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr12_+_69230931 | 0.08 |
ENSMUST00000060579.10
|
Mgat2
|
mannoside acetylglucosaminyltransferase 2 |
| chr10_-_121933276 | 0.08 |
ENSMUST00000140299.3
|
Rxylt1
|
ribitol xylosyltransferase 1 |
| chr5_+_110324506 | 0.08 |
ENSMUST00000112512.8
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr3_-_84489783 | 0.08 |
ENSMUST00000107687.9
ENSMUST00000098990.10 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr1_+_87254729 | 0.07 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr2_-_168072493 | 0.07 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr16_-_10952465 | 0.07 |
ENSMUST00000118362.8
ENSMUST00000118679.2 ENSMUST00000038424.14 |
Txndc11
|
thioredoxin domain containing 11 |
| chr12_-_36092475 | 0.07 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr19_+_8719033 | 0.07 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
| chr8_+_71819534 | 0.07 |
ENSMUST00000110054.8
ENSMUST00000139541.8 |
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr7_-_126975199 | 0.05 |
ENSMUST00000067425.6
|
Zfp747
|
zinc finger protein 747 |
| chr17_-_26080429 | 0.05 |
ENSMUST00000079461.15
ENSMUST00000176923.9 |
Wdr90
|
WD repeat domain 90 |
| chr8_+_71819854 | 0.04 |
ENSMUST00000019169.8
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr3_+_89153704 | 0.04 |
ENSMUST00000168900.3
|
Krtcap2
|
keratinocyte associated protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 2.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.6 | 2.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 2.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.5 | 3.8 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.4 | 4.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 2.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 5.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 1.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 0.9 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 1.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 1.2 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 0.7 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.2 | 0.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.2 | 0.6 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.2 | 1.7 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 1.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 2.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 1.2 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 3.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 1.0 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 6.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.4 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.3 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 2.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 1.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.8 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 4.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 1.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.5 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.4 | 1.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 4.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 5.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 4.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 1.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 28.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.6 | 2.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 2.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 2.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 3.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 2.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 4.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 2.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 3.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.5 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 6.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 1.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 11.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |