Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
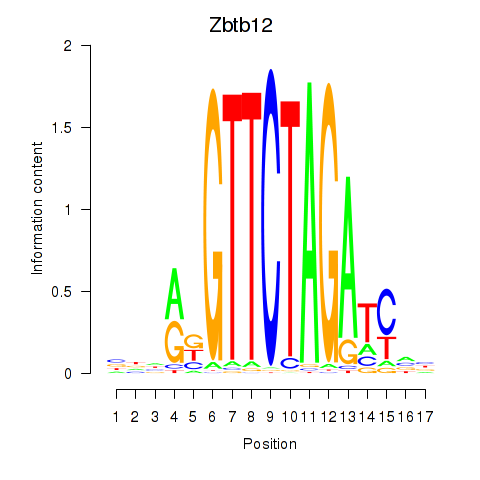
Results for Zbtb12
Z-value: 0.42
Transcription factors associated with Zbtb12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb12
|
ENSMUSG00000049823.10 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | mm39_v1_chr17_+_35113490_35113554 | -0.30 | 7.3e-02 | Click! |
Activity profile of Zbtb12 motif
Sorted Z-values of Zbtb12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_25239229 | 2.30 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr7_-_25176959 | 1.36 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr15_-_60793115 | 1.30 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr11_-_70590923 | 1.20 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr19_-_8196196 | 1.20 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr17_+_37253802 | 0.92 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr2_+_43445333 | 0.88 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr2_+_30254239 | 0.87 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr11_+_70591299 | 0.80 |
ENSMUST00000152618.9
ENSMUST00000102554.8 ENSMUST00000094499.11 ENSMUST00000072187.12 ENSMUST00000137119.3 |
Kif1c
|
kinesin family member 1C |
| chr6_+_90439544 | 0.75 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439596 | 0.75 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr2_+_43445359 | 0.73 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr7_-_48493388 | 0.67 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr17_+_79934096 | 0.62 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr15_+_76544058 | 0.56 |
ENSMUST00000230451.2
|
Kifc2
|
kinesin family member C2 |
| chr15_-_76543889 | 0.54 |
ENSMUST00000231152.2
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr16_-_94657531 | 0.45 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr4_-_45489794 | 0.44 |
ENSMUST00000146236.8
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr9_-_35030479 | 0.44 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr10_+_62896492 | 0.41 |
ENSMUST00000219687.2
ENSMUST00000219045.2 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr14_+_67470884 | 0.40 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr6_+_37877413 | 0.38 |
ENSMUST00000120238.2
|
Trim24
|
tripartite motif-containing 24 |
| chr11_-_50806962 | 0.37 |
ENSMUST00000116378.8
ENSMUST00000109128.8 |
Zfp2
|
zinc finger protein 2 |
| chr7_+_100966289 | 0.36 |
ENSMUST00000163799.9
ENSMUST00000164479.9 |
Stard10
|
START domain containing 10 |
| chr13_+_9326513 | 0.35 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr19_+_46561801 | 0.34 |
ENSMUST00000026011.8
|
Sfxn2
|
sideroflexin 2 |
| chr4_-_43669141 | 0.34 |
ENSMUST00000056474.7
|
Fam221b
|
family with sequence similarity 221, member B |
| chr19_-_57170725 | 0.34 |
ENSMUST00000133369.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr9_-_58448224 | 0.33 |
ENSMUST00000039788.11
|
Cd276
|
CD276 antigen |
| chr11_+_114743044 | 0.32 |
ENSMUST00000142262.2
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_108271990 | 0.32 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr4_+_43669266 | 0.30 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr17_+_37253916 | 0.30 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr6_-_124791259 | 0.28 |
ENSMUST00000172132.10
ENSMUST00000239432.2 |
Tpi1
|
triosephosphate isomerase 1 |
| chr11_-_118021460 | 0.25 |
ENSMUST00000132685.9
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr4_-_119031050 | 0.24 |
ENSMUST00000052715.10
ENSMUST00000179290.8 ENSMUST00000154226.2 |
Zfp691
|
zinc finger protein 691 |
| chr17_-_28779678 | 0.24 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr3_-_131138541 | 0.23 |
ENSMUST00000090246.5
ENSMUST00000126569.2 |
Sgms2
|
sphingomyelin synthase 2 |
| chr9_+_36604516 | 0.23 |
ENSMUST00000034620.5
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr6_-_24528012 | 0.22 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr14_-_70449438 | 0.21 |
ENSMUST00000227929.2
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr15_-_82238432 | 0.20 |
ENSMUST00000230494.2
ENSMUST00000023085.7 |
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr8_-_107007596 | 0.20 |
ENSMUST00000212896.2
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr18_+_70605691 | 0.20 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr18_+_70605476 | 0.20 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr15_+_102412157 | 0.20 |
ENSMUST00000096145.5
|
Gm10337
|
predicted gene 10337 |
| chr18_+_70605722 | 0.19 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr13_-_104365310 | 0.19 |
ENSMUST00000022226.6
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr4_-_138351313 | 0.19 |
ENSMUST00000030533.12
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr6_+_108041998 | 0.18 |
ENSMUST00000049246.7
|
Setmar
|
SET domain without mariner transposase fusion |
| chr4_+_119396913 | 0.18 |
ENSMUST00000137560.8
|
Foxj3
|
forkhead box J3 |
| chr15_-_88838605 | 0.18 |
ENSMUST00000109371.8
|
Ttll8
|
tubulin tyrosine ligase-like family, member 8 |
| chr18_+_70605630 | 0.17 |
ENSMUST00000168249.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr9_+_38407097 | 0.17 |
ENSMUST00000214003.3
ENSMUST00000214264.3 |
Olfr907
|
olfactory receptor 907 |
| chr11_+_97697128 | 0.17 |
ENSMUST00000138919.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr10_-_85847697 | 0.16 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr3_+_121243395 | 0.16 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr7_+_140427711 | 0.16 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr7_+_140427729 | 0.16 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr10_-_75658355 | 0.15 |
ENSMUST00000160211.2
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr11_+_98337655 | 0.15 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr13_+_76246853 | 0.15 |
ENSMUST00000091466.4
ENSMUST00000224386.2 |
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr2_-_150531280 | 0.15 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr11_+_102775991 | 0.14 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr10_+_63293284 | 0.14 |
ENSMUST00000105440.8
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr17_-_32408431 | 0.13 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr1_-_161807205 | 0.13 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr11_+_100332709 | 0.13 |
ENSMUST00000001599.4
ENSMUST00000107395.3 |
Klhl10
|
kelch-like 10 |
| chr13_-_92620507 | 0.13 |
ENSMUST00000040106.9
|
Fam151b
|
family with sequence similarity 151, member B |
| chr7_+_99825886 | 0.13 |
ENSMUST00000178946.9
|
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr14_-_52728503 | 0.13 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
| chr8_+_71156071 | 0.13 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr14_+_8555284 | 0.12 |
ENSMUST00000144914.3
|
Gm281
|
predicted gene 281 |
| chr17_-_15596230 | 0.12 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr1_-_75208734 | 0.12 |
ENSMUST00000180101.3
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr6_-_143892814 | 0.11 |
ENSMUST00000124233.7
ENSMUST00000144289.4 ENSMUST00000111748.8 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr12_-_81615248 | 0.11 |
ENSMUST00000008582.4
|
Adam21
|
a disintegrin and metallopeptidase domain 21 |
| chr15_-_99372575 | 0.11 |
ENSMUST00000040313.6
|
Bcdin3d
|
BCDIN3 domain containing |
| chr6_+_24528143 | 0.11 |
ENSMUST00000031696.10
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr1_+_69866145 | 0.10 |
ENSMUST00000065425.12
ENSMUST00000113940.4 |
Spag16
|
sperm associated antigen 16 |
| chr8_-_43981143 | 0.10 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chr6_+_136528155 | 0.10 |
ENSMUST00000186742.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_102420428 | 0.10 |
ENSMUST00000213432.2
|
Olfr561
|
olfactory receptor 561 |
| chr5_+_87955941 | 0.10 |
ENSMUST00000072539.12
ENSMUST00000113279.8 ENSMUST00000101057.8 ENSMUST00000197301.2 |
Csn1s2b
|
casein alpha s2-like B |
| chr3_-_153610528 | 0.10 |
ENSMUST00000190449.2
|
Msh4
|
mutS homolog 4 |
| chr3_-_153610368 | 0.09 |
ENSMUST00000188338.7
|
Msh4
|
mutS homolog 4 |
| chr9_-_40366966 | 0.09 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr14_+_79718604 | 0.09 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr9_-_122673080 | 0.09 |
ENSMUST00000203176.3
ENSMUST00000203656.3 ENSMUST00000204619.2 |
Gm35549
|
predicted gene, 35549 |
| chr10_-_5144699 | 0.09 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr7_-_35285001 | 0.09 |
ENSMUST00000069912.6
|
Rgs9bp
|
regulator of G-protein signalling 9 binding protein |
| chr2_-_111942878 | 0.08 |
ENSMUST00000217078.2
|
Olfr1315-ps1
|
olfactory receptor 1315, pseudogene 1 |
| chr5_+_96104775 | 0.08 |
ENSMUST00000023840.7
|
Cxcl13
|
chemokine (C-X-C motif) ligand 13 |
| chr5_+_11234174 | 0.08 |
ENSMUST00000168407.3
|
Gm5861
|
predicted gene 5861 |
| chr15_+_89383799 | 0.08 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_+_5283606 | 0.08 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr12_+_111540920 | 0.08 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr3_-_152232389 | 0.08 |
ENSMUST00000200062.2
|
Ak5
|
adenylate kinase 5 |
| chr5_-_142515792 | 0.08 |
ENSMUST00000099400.3
|
Papolb
|
poly (A) polymerase beta (testis specific) |
| chr11_-_59118988 | 0.08 |
ENSMUST00000163300.8
ENSMUST00000061242.8 |
Arf1
|
ADP-ribosylation factor 1 |
| chr15_-_76259126 | 0.08 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr14_+_26789345 | 0.07 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr14_+_26414422 | 0.07 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr17_+_83658354 | 0.07 |
ENSMUST00000096766.12
ENSMUST00000049503.10 ENSMUST00000112363.10 ENSMUST00000234460.2 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr7_+_65693350 | 0.07 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
| chr18_-_20995980 | 0.07 |
ENSMUST00000224530.2
|
Trappc8
|
trafficking protein particle complex 8 |
| chrX_+_106836189 | 0.06 |
ENSMUST00000101292.9
|
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr5_-_22070602 | 0.06 |
ENSMUST00000030878.8
|
Slc26a5
|
solute carrier family 26, member 5 |
| chrX_-_36167148 | 0.06 |
ENSMUST00000239259.2
|
Nkrf
|
NF-kappaB repressing factor |
| chr14_+_54082691 | 0.06 |
ENSMUST00000103674.6
|
Trav19
|
T cell receptor alpha variable 19 |
| chr15_-_98796373 | 0.05 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr3_+_5283577 | 0.04 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr11_+_104983022 | 0.04 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr8_+_41205245 | 0.04 |
ENSMUST00000096663.5
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2) |
| chrX_+_93768175 | 0.04 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr17_-_57181420 | 0.04 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr4_-_133360749 | 0.03 |
ENSMUST00000084238.5
|
Zdhhc18
|
zinc finger, DHHC domain containing 18 |
| chr10_-_70428611 | 0.03 |
ENSMUST00000162251.8
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr19_+_6130059 | 0.03 |
ENSMUST00000149347.8
ENSMUST00000143303.2 |
Tmem262
|
transmembrane protein 262 |
| chr13_-_14787602 | 0.03 |
ENSMUST00000220621.2
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr3_-_84063067 | 0.03 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr12_-_101924407 | 0.02 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr9_+_37278647 | 0.02 |
ENSMUST00000051839.9
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr11_+_69881885 | 0.02 |
ENSMUST00000018711.15
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chrX_+_41591476 | 0.02 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr17_+_27248233 | 0.02 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr2_+_129854256 | 0.01 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr4_+_32983417 | 0.01 |
ENSMUST00000084747.6
|
Rragd
|
Ras-related GTP binding D |
| chr9_-_78389006 | 0.01 |
ENSMUST00000042235.15
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr5_-_35836761 | 0.00 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr5_-_121641461 | 0.00 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
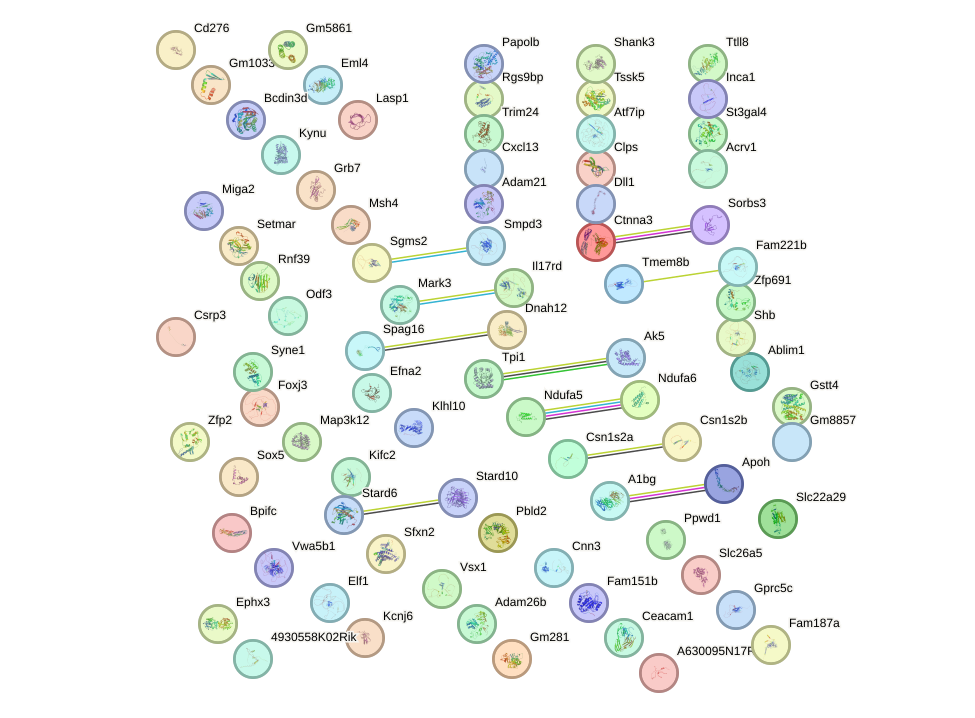
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 1.4 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 0.2 | 0.7 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.2 | 2.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 1.5 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 1.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 3.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 1.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |