Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
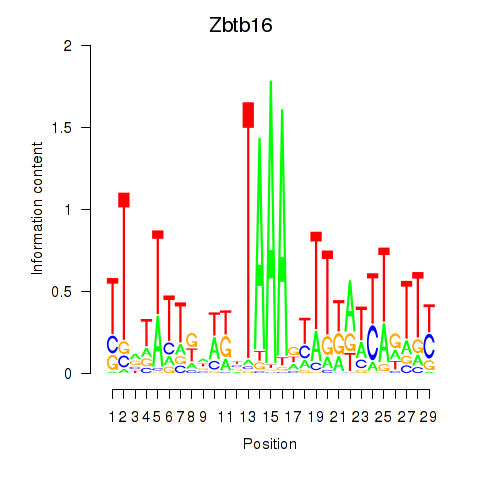
Results for Zbtb16
Z-value: 0.45
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSMUSG00000066687.6 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | mm39_v1_chr9_-_48747232_48747262 | -0.32 | 5.8e-02 | Click! |
Activity profile of Zbtb16 motif
Sorted Z-values of Zbtb16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_44300876 | 1.12 |
ENSMUST00000045607.12
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr5_-_148988110 | 0.92 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr11_+_23615612 | 0.90 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr7_-_133304244 | 0.87 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr4_+_109272828 | 0.86 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr7_-_45173193 | 0.83 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr4_-_117035922 | 0.78 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr1_+_127657142 | 0.77 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr1_-_82746169 | 0.76 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr9_+_96140781 | 0.72 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr4_+_33310306 | 0.72 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr9_+_96141299 | 0.71 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr5_-_65492940 | 0.71 |
ENSMUST00000203471.3
ENSMUST00000172732.8 ENSMUST00000204965.3 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr4_+_123176570 | 0.69 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr9_+_96141317 | 0.67 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr14_-_66071412 | 0.65 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr4_+_15957923 | 0.64 |
ENSMUST00000029879.15
ENSMUST00000149069.2 |
Nbn
|
nibrin |
| chr4_-_119177614 | 0.64 |
ENSMUST00000147077.8
ENSMUST00000056458.14 ENSMUST00000106321.9 ENSMUST00000106319.8 ENSMUST00000106317.2 ENSMUST00000106318.8 |
Ppih
|
peptidyl prolyl isomerase H |
| chr9_+_119939414 | 0.64 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr7_-_115445352 | 0.62 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_71142305 | 0.61 |
ENSMUST00000027393.8
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr15_-_57982705 | 0.59 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr9_+_96140750 | 0.59 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr17_-_27816151 | 0.56 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr7_-_115445315 | 0.56 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_23436620 | 0.54 |
ENSMUST00000188677.2
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr4_-_19570073 | 0.53 |
ENSMUST00000029885.5
|
Cpne3
|
copine III |
| chr11_-_99328969 | 0.53 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr5_-_65492907 | 0.48 |
ENSMUST00000203581.3
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr5_-_72325482 | 0.48 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chrX_-_135104589 | 0.46 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr10_-_93727003 | 0.46 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr15_-_83479312 | 0.45 |
ENSMUST00000016901.5
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr7_+_27259895 | 0.45 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr5_+_110478558 | 0.44 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr15_-_96597610 | 0.44 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr12_+_55286111 | 0.42 |
ENSMUST00000164243.2
|
Srp54c
|
signal recognition particle 54C |
| chr11_-_68899248 | 0.41 |
ENSMUST00000021282.12
|
Pfas
|
phosphoribosylformylglycinamidine synthase (FGAR amidotransferase) |
| chr19_+_53128901 | 0.41 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr11_+_68936457 | 0.40 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr11_-_86248395 | 0.40 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr19_-_8382424 | 0.40 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr10_-_23112973 | 0.40 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr7_+_101520843 | 0.39 |
ENSMUST00000210984.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr19_+_53298906 | 0.39 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr2_-_145776934 | 0.38 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr19_-_8196196 | 0.37 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr15_+_54274151 | 0.37 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr2_+_80469142 | 0.37 |
ENSMUST00000028382.13
ENSMUST00000124377.2 |
Nup35
|
nucleoporin 35 |
| chr17_+_26471870 | 0.37 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr17_+_44499451 | 0.37 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr3_+_32760447 | 0.36 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chr10_-_25172953 | 0.36 |
ENSMUST00000177124.2
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr11_+_43324547 | 0.35 |
ENSMUST00000126128.8
ENSMUST00000151880.8 ENSMUST00000020681.10 |
Slu7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr5_-_121665249 | 0.35 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chrX_-_133012600 | 0.35 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr17_+_26471889 | 0.34 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr4_-_102883905 | 0.34 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr19_+_53128861 | 0.34 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr10_-_116385007 | 0.34 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr18_-_67582191 | 0.33 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr4_-_88595161 | 0.33 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr18_-_35795233 | 0.33 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr16_-_75706161 | 0.32 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr4_-_135780660 | 0.32 |
ENSMUST00000102536.11
|
Rpl11
|
ribosomal protein L11 |
| chr2_+_132532040 | 0.32 |
ENSMUST00000148271.8
ENSMUST00000110132.3 |
Shld1
|
shieldin complex subunit 1 |
| chr1_+_179936757 | 0.32 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr9_-_122695071 | 0.32 |
ENSMUST00000216063.2
|
Zfp445
|
zinc finger protein 445 |
| chr1_-_120197979 | 0.32 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr11_-_93856783 | 0.31 |
ENSMUST00000021220.10
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr6_-_118396321 | 0.31 |
ENSMUST00000032237.8
|
Bms1
|
BMS1, ribosome biogenesis factor |
| chrX_-_7054952 | 0.30 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr9_+_62754252 | 0.30 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr5_-_130284366 | 0.29 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr13_-_8921732 | 0.29 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr5_+_143534455 | 0.29 |
ENSMUST00000169329.8
ENSMUST00000067145.12 ENSMUST00000119488.2 ENSMUST00000118121.2 ENSMUST00000200267.2 ENSMUST00000196487.2 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr13_+_54225828 | 0.29 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr16_+_36097505 | 0.29 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr16_+_36097313 | 0.28 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr11_+_87685032 | 0.28 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr17_+_46807637 | 0.27 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr1_-_128345089 | 0.27 |
ENSMUST00000027602.15
ENSMUST00000064309.9 |
Dars
|
aspartyl-tRNA synthetase |
| chr7_+_103620359 | 0.27 |
ENSMUST00000209473.4
|
Olfr635
|
olfactory receptor 635 |
| chr2_-_165700055 | 0.26 |
ENSMUST00000109266.11
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr17_+_43327412 | 0.26 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr4_-_94444975 | 0.26 |
ENSMUST00000030313.9
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr12_-_75678092 | 0.26 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr19_-_32717166 | 0.26 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr9_-_44679136 | 0.25 |
ENSMUST00000034607.10
|
Arcn1
|
archain 1 |
| chr1_-_120198804 | 0.25 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr9_-_50528727 | 0.25 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr19_+_39102342 | 0.24 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr17_-_65901946 | 0.24 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr14_-_26184767 | 0.24 |
ENSMUST00000146438.2
|
Slmap
|
sarcolemma associated protein |
| chr9_+_20209828 | 0.23 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr16_-_20972750 | 0.23 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr6_+_37877413 | 0.23 |
ENSMUST00000120238.2
|
Trim24
|
tripartite motif-containing 24 |
| chr9_+_54493618 | 0.23 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr2_-_25517945 | 0.22 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr15_-_98560739 | 0.22 |
ENSMUST00000162384.2
ENSMUST00000003450.15 |
Ddx23
|
DEAD box helicase 23 |
| chr11_-_48717482 | 0.22 |
ENSMUST00000104959.2
|
Gm12184
|
predicted gene 12184 |
| chr5_-_135423353 | 0.22 |
ENSMUST00000111171.6
|
Pom121
|
nuclear pore membrane protein 121 |
| chr9_+_20799471 | 0.22 |
ENSMUST00000004203.6
|
Ppan
|
peter pan homolog |
| chr10_-_127358231 | 0.22 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_-_168607166 | 0.21 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr19_-_8691797 | 0.21 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr5_+_149601688 | 0.21 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr13_-_21586858 | 0.21 |
ENSMUST00000117721.8
ENSMUST00000070785.16 ENSMUST00000116433.2 ENSMUST00000223831.2 ENSMUST00000116434.11 ENSMUST00000224820.2 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr10_+_22236451 | 0.21 |
ENSMUST00000182677.8
|
Raet1d
|
retinoic acid early transcript delta |
| chr2_-_37537224 | 0.21 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr1_+_128079543 | 0.21 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr11_-_17903861 | 0.21 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr19_-_8763771 | 0.20 |
ENSMUST00000176496.8
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chrX_-_162859429 | 0.20 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr10_-_127358300 | 0.20 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_+_125994050 | 0.20 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr2_+_15054206 | 0.20 |
ENSMUST00000017562.13
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr4_+_129181407 | 0.20 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr3_-_41696906 | 0.20 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr19_-_8691460 | 0.19 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr17_+_36190662 | 0.19 |
ENSMUST00000025292.15
|
Dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_-_101308441 | 0.19 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr17_-_53846438 | 0.19 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr6_+_91855015 | 0.19 |
ENSMUST00000037783.7
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr19_-_5452521 | 0.18 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr13_-_22375311 | 0.18 |
ENSMUST00000238109.2
|
Vmn1r192
|
vomeronasal 1 receptor 192 |
| chr15_+_79555272 | 0.18 |
ENSMUST00000127292.2
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chr18_-_35795175 | 0.18 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr17_+_46421908 | 0.18 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr19_-_32717138 | 0.18 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr7_-_16121682 | 0.18 |
ENSMUST00000094815.5
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr7_+_75292971 | 0.17 |
ENSMUST00000207998.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_73382303 | 0.17 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr14_-_73785682 | 0.17 |
ENSMUST00000043813.3
|
Nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr9_-_122673080 | 0.17 |
ENSMUST00000203176.3
ENSMUST00000203656.3 ENSMUST00000204619.2 |
Gm35549
|
predicted gene, 35549 |
| chr6_+_136495784 | 0.17 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr11_-_73348284 | 0.17 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr6_+_70332836 | 0.16 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr13_+_94925382 | 0.16 |
ENSMUST00000046644.7
|
Tbca
|
tubulin cofactor A |
| chr1_-_171362852 | 0.16 |
ENSMUST00000043094.13
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr5_-_18093739 | 0.16 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr11_-_115590318 | 0.16 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chrX_-_9335525 | 0.16 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr1_+_107439145 | 0.16 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr7_-_133384449 | 0.16 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_-_58912678 | 0.16 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr9_-_71803354 | 0.15 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr12_-_91556761 | 0.15 |
ENSMUST00000021345.14
|
Gtf2a1
|
general transcription factor II A, 1 |
| chr4_+_32623985 | 0.15 |
ENSMUST00000108178.2
|
Casp8ap2
|
caspase 8 associated protein 2 |
| chr10_+_123099945 | 0.15 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr5_-_134258435 | 0.15 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr11_-_106889291 | 0.15 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr2_-_169973076 | 0.15 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr6_+_56962280 | 0.15 |
ENSMUST00000164307.2
|
Vmn1r5
|
vomeronasal 1 receptor 5 |
| chr11_-_77678573 | 0.15 |
ENSMUST00000092883.3
|
Gm10277
|
predicted gene 10277 |
| chr2_-_15054065 | 0.14 |
ENSMUST00000028034.15
ENSMUST00000114713.2 |
Nsun6
|
NOL1/NOP2/Sun domain family member 6 |
| chr10_-_81200680 | 0.14 |
ENSMUST00000131736.8
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr2_-_36979237 | 0.14 |
ENSMUST00000216663.2
ENSMUST00000214969.2 |
Olfr361
|
olfactory receptor 361 |
| chr5_-_23821523 | 0.14 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_-_125090540 | 0.14 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr6_+_57234937 | 0.14 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr18_+_38809771 | 0.14 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr19_+_4008645 | 0.13 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr5_-_105130522 | 0.13 |
ENSMUST00000031239.13
|
Abcg3
|
ATP binding cassette subfamily G member 3 |
| chr7_-_125090757 | 0.13 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr1_-_63153414 | 0.13 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr9_-_65815958 | 0.13 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr2_+_89865681 | 0.13 |
ENSMUST00000214855.2
|
Olfr1265
|
olfactory receptor 1265 |
| chr4_-_126094910 | 0.13 |
ENSMUST00000136157.8
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr14_+_51366306 | 0.13 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chrX_-_156381652 | 0.13 |
ENSMUST00000149249.2
ENSMUST00000058098.15 |
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr14_+_53153256 | 0.12 |
ENSMUST00000197007.2
ENSMUST00000103593.3 |
Trav12d-2
|
T cell receptor alpha variable 12D-2 |
| chr3_-_151960992 | 0.12 |
ENSMUST00000198750.5
|
Nexn
|
nexilin |
| chr1_+_109911467 | 0.12 |
ENSMUST00000172005.8
|
Cdh7
|
cadherin 7, type 2 |
| chr7_-_104050027 | 0.12 |
ENSMUST00000106828.3
|
Trim30c
|
tripartite motif-containing 30C |
| chr3_-_146475974 | 0.12 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr19_-_29344694 | 0.11 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr1_-_124773767 | 0.11 |
ENSMUST00000239072.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr9_+_38766356 | 0.11 |
ENSMUST00000104874.3
|
Olfr26
|
olfactory receptor 26 |
| chr18_+_13088878 | 0.11 |
ENSMUST00000234763.2
|
Impact
|
impact, RWD domain protein |
| chr13_+_55300453 | 0.11 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr5_-_143123955 | 0.11 |
ENSMUST00000218872.3
|
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr11_-_78074377 | 0.11 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr9_+_104930438 | 0.11 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr2_+_85545763 | 0.11 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr7_-_109330915 | 0.10 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr14_+_53194239 | 0.10 |
ENSMUST00000199800.2
|
Trav15d-2-dv6d-2
|
T cell receptor alpha variable 15D-2-DV6D-2 |
| chr7_-_29898236 | 0.10 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr8_+_104977493 | 0.10 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr17_-_71305003 | 0.10 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr9_+_39903409 | 0.10 |
ENSMUST00000217600.2
|
Olfr978
|
olfactory receptor 978 |
| chr2_-_87838612 | 0.10 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr18_+_37102969 | 0.10 |
ENSMUST00000194544.2
|
Gm37013
|
predicted gene, 37013 |
| chr10_+_33780993 | 0.10 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr7_+_51537645 | 0.10 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr18_-_3299452 | 0.10 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr15_-_36140539 | 0.10 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr16_+_58967409 | 0.10 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr17_+_24072493 | 0.10 |
ENSMUST00000061725.8
|
Prss32
|
protease, serine 32 |
| chr18_-_3299536 | 0.10 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
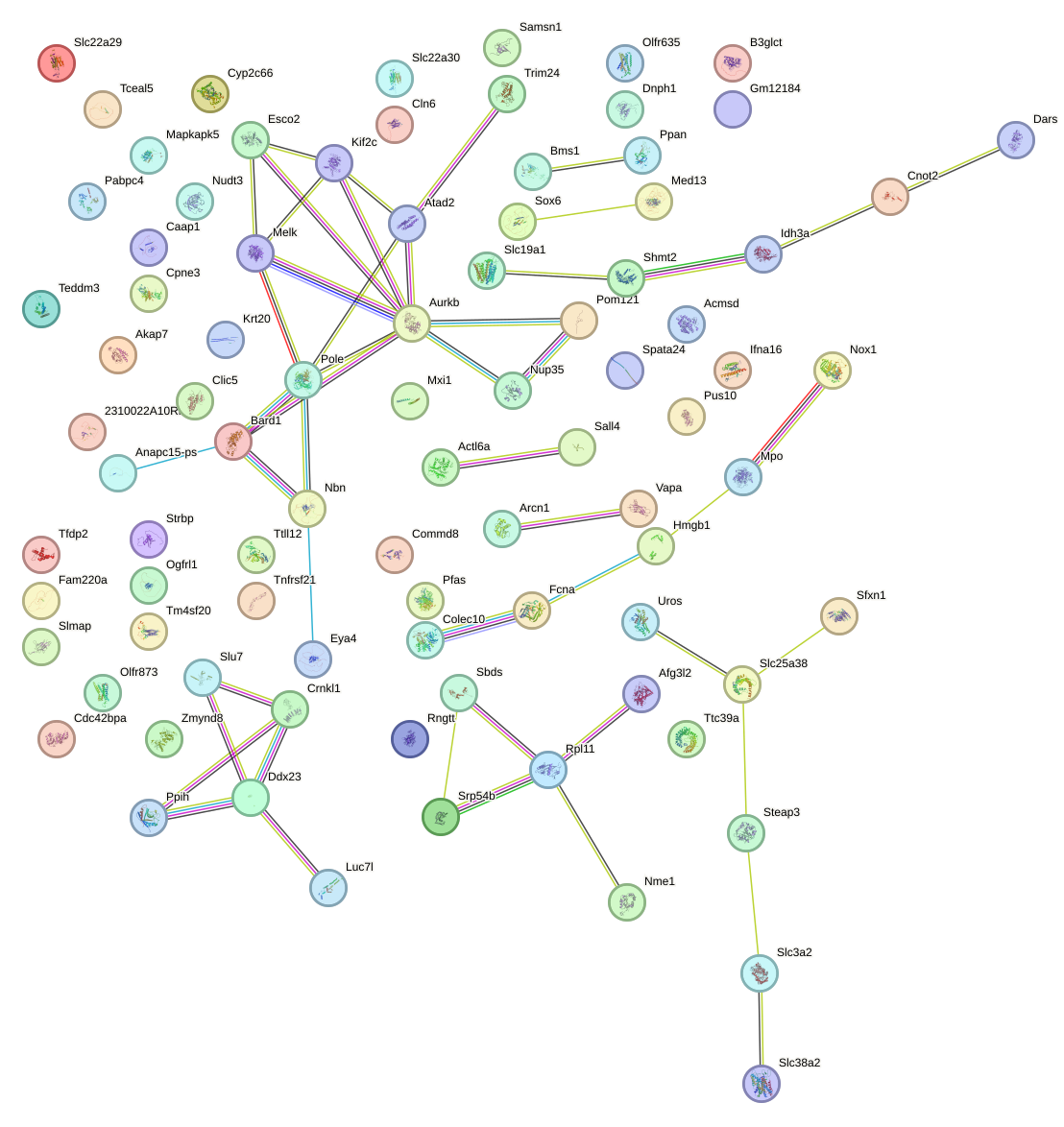
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb16
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.3 | 0.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.3 | 0.8 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.2 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 0.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.6 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 1.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.3 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.6 | GO:0085020 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.3 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.4 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.1 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 1.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.1 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.6 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |