Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
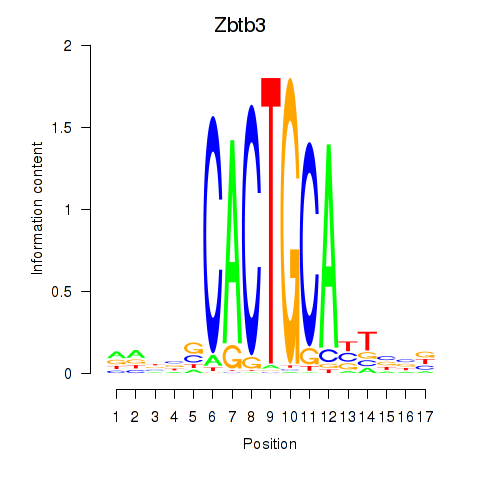
Results for Zbtb3
Z-value: 0.54
Transcription factors associated with Zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb3
|
ENSMUSG00000071661.8 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | mm39_v1_chr19_+_8779903_8779922 | -0.82 | 8.3e-10 | Click! |
Activity profile of Zbtb3 motif
Sorted Z-values of Zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_8382424 | 4.48 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr7_-_114162125 | 3.73 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr1_-_121255448 | 3.62 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255753 | 3.58 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121255400 | 3.40 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr9_+_46180362 | 3.29 |
ENSMUST00000214202.2
ENSMUST00000215458.2 ENSMUST00000215187.2 ENSMUST00000213878.2 ENSMUST00000034584.4 |
Apoa5
|
apolipoprotein A-V |
| chr1_-_121255503 | 2.72 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr16_+_22739028 | 2.67 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr16_+_22739191 | 2.59 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr16_+_22738987 | 2.57 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr19_-_8109346 | 1.88 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr18_-_16942289 | 1.88 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr1_-_173195236 | 1.87 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr10_-_53951796 | 1.74 |
ENSMUST00000105470.9
|
Man1a
|
mannosidase 1, alpha |
| chr10_-_53951825 | 1.67 |
ENSMUST00000003843.16
|
Man1a
|
mannosidase 1, alpha |
| chr8_-_3517617 | 1.67 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr19_-_58444336 | 1.56 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr10_+_61531282 | 1.55 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr9_+_92157655 | 1.51 |
ENSMUST00000034932.14
ENSMUST00000180154.8 |
Plscr2
|
phospholipid scramblase 2 |
| chr1_-_133849131 | 1.51 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr14_+_53995108 | 1.48 |
ENSMUST00000184905.2
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr11_-_120622770 | 1.35 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr19_-_34855278 | 1.25 |
ENSMUST00000112460.3
|
Pank1
|
pantothenate kinase 1 |
| chr11_+_87482971 | 1.21 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr9_+_21746785 | 1.19 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr19_-_34855242 | 1.17 |
ENSMUST00000238065.2
|
Pank1
|
pantothenate kinase 1 |
| chr2_-_127634387 | 1.16 |
ENSMUST00000135091.2
|
Mtln
|
mitoregulin |
| chr5_-_90371816 | 1.16 |
ENSMUST00000118816.6
ENSMUST00000048363.10 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chr19_-_59064501 | 1.15 |
ENSMUST00000163821.3
ENSMUST00000047511.15 |
Shtn1
|
shootin 1 |
| chr6_-_86503178 | 1.14 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr10_+_128104525 | 1.06 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr1_-_162687369 | 1.03 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr1_-_162687254 | 0.97 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_+_80447389 | 0.94 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr14_+_53399856 | 0.93 |
ENSMUST00000198359.2
|
Trav13n-1
|
T cell receptor alpha variable 13N-1 |
| chr8_-_123980908 | 0.91 |
ENSMUST00000122363.8
|
Vps9d1
|
VPS9 domain containing 1 |
| chr19_-_21630143 | 0.89 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr4_-_150093435 | 0.88 |
ENSMUST00000030830.4
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr19_+_25214322 | 0.88 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr14_+_36789999 | 0.82 |
ENSMUST00000057176.5
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr9_+_78099229 | 0.81 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr15_+_103181311 | 0.80 |
ENSMUST00000100162.5
ENSMUST00000230893.2 |
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr14_+_53872276 | 0.78 |
ENSMUST00000103658.4
|
Trav13-2
|
T cell receptor alpha variable 13-2 |
| chr11_-_72380730 | 0.77 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr14_+_53574579 | 0.75 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr9_+_44410417 | 0.74 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr14_+_53601293 | 0.67 |
ENSMUST00000103634.3
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr11_+_105956867 | 0.67 |
ENSMUST00000002048.8
|
Taco1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr9_+_66853343 | 0.64 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr4_-_49473904 | 0.63 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr3_+_79793237 | 0.63 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr14_-_19261196 | 0.58 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr8_+_65399831 | 0.57 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr9_+_43980718 | 0.52 |
ENSMUST00000114830.9
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr6_-_147165623 | 0.50 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chr14_+_53270305 | 0.48 |
ENSMUST00000179512.3
|
Trav13d-3
|
T cell receptor alpha variable 13D-3 |
| chr14_+_53491504 | 0.48 |
ENSMUST00000103622.3
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr12_+_64964674 | 0.48 |
ENSMUST00000058135.6
ENSMUST00000220993.2 |
Gm527
|
predicted gene 527 |
| chr14_-_50919102 | 0.47 |
ENSMUST00000078075.5
|
Olfr747
|
olfactory receptor 747 |
| chr4_+_40269563 | 0.46 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr14_+_53491249 | 0.46 |
ENSMUST00000196941.2
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr16_+_34470290 | 0.46 |
ENSMUST00000148562.8
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr18_+_37440497 | 0.45 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr18_-_3299452 | 0.44 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr1_-_172085977 | 0.43 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr7_+_97049210 | 0.43 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr14_+_53180476 | 0.43 |
ENSMUST00000103596.3
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr16_-_59459745 | 0.43 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr7_+_101555111 | 0.41 |
ENSMUST00000033131.12
ENSMUST00000193465.2 |
Lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr16_+_20408886 | 0.41 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr2_+_119378178 | 0.40 |
ENSMUST00000014221.13
ENSMUST00000119172.2 |
Chp1
|
calcineurin-like EF hand protein 1 |
| chr7_+_28466658 | 0.39 |
ENSMUST00000155327.8
|
Sirt2
|
sirtuin 2 |
| chr11_-_3724706 | 0.39 |
ENSMUST00000155197.2
|
Osbp2
|
oxysterol binding protein 2 |
| chr2_-_51862941 | 0.39 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr11_+_92989229 | 0.38 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr1_+_181952302 | 0.38 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr5_-_45796857 | 0.38 |
ENSMUST00000016023.9
|
Fam184b
|
family with sequence similarity 184, member B |
| chr2_-_119378108 | 0.37 |
ENSMUST00000060009.9
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr5_-_39034039 | 0.36 |
ENSMUST00000169819.5
ENSMUST00000171633.5 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr9_+_92157799 | 0.35 |
ENSMUST00000126911.2
|
Plscr2
|
phospholipid scramblase 2 |
| chr9_+_30338329 | 0.35 |
ENSMUST00000164099.3
|
Snx19
|
sorting nexin 19 |
| chr18_+_44467133 | 0.34 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr10_+_76089674 | 0.34 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr2_+_140012560 | 0.33 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr7_-_30826376 | 0.33 |
ENSMUST00000098548.8
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr19_-_58781709 | 0.32 |
ENSMUST00000166692.2
|
1700019N19Rik
|
RIKEN cDNA 1700019N19 gene |
| chr9_+_107458495 | 0.32 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr7_-_62862261 | 0.32 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr9_-_89505178 | 0.32 |
ENSMUST00000044491.13
|
Minar1
|
membrane integral NOTCH2 associated receptor 1 |
| chr7_+_138448308 | 0.31 |
ENSMUST00000155672.8
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr2_-_119377675 | 0.30 |
ENSMUST00000133668.2
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr13_-_95359543 | 0.29 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr16_-_56616186 | 0.29 |
ENSMUST00000023437.5
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr13_-_96678844 | 0.29 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr14_+_53539493 | 0.28 |
ENSMUST00000103627.3
|
Trav4n-4
|
T cell receptor alpha variable 4N-4 |
| chr11_+_58868919 | 0.28 |
ENSMUST00000108809.8
ENSMUST00000108810.10 ENSMUST00000093061.7 |
Trim11
|
tripartite motif-containing 11 |
| chr17_-_57064516 | 0.28 |
ENSMUST00000044752.6
|
Nrtn
|
neurturin |
| chr11_-_5691117 | 0.27 |
ENSMUST00000140922.2
ENSMUST00000093362.12 |
Urgcp
|
upregulator of cell proliferation |
| chr10_-_62723103 | 0.27 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr11_-_99979052 | 0.26 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr16_-_59459486 | 0.26 |
ENSMUST00000149797.3
ENSMUST00000023405.16 |
Arl6
|
ADP-ribosylation factor-like 6 |
| chr6_+_18170686 | 0.26 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr11_-_58504307 | 0.26 |
ENSMUST00000048801.8
|
Lypd8l
|
LY6/PLAUR domain containing 8 like |
| chr2_-_51863203 | 0.25 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr7_+_18758289 | 0.25 |
ENSMUST00000023882.14
ENSMUST00000153976.8 |
Sympk
|
symplekin |
| chr14_+_53180221 | 0.25 |
ENSMUST00000197954.2
|
Trav13d-2
|
T cell receptor alpha variable 13D-2 |
| chr2_+_113158053 | 0.25 |
ENSMUST00000099576.9
|
Fmn1
|
formin 1 |
| chr6_-_40590244 | 0.25 |
ENSMUST00000076565.3
|
Tas2r138
|
taste receptor, type 2, member 138 |
| chr9_+_32027335 | 0.25 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_+_34380738 | 0.25 |
ENSMUST00000066133.7
|
Apc
|
APC, WNT signaling pathway regulator |
| chr14_+_53782432 | 0.24 |
ENSMUST00000103651.4
|
Trav13-1
|
T cell receptor alpha variable 13-1 |
| chr7_-_121700958 | 0.24 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr1_+_156138286 | 0.24 |
ENSMUST00000027896.10
|
Nphs2
|
nephrosis 2, podocin |
| chr2_+_113158100 | 0.24 |
ENSMUST00000102547.10
|
Fmn1
|
formin 1 |
| chr7_+_141276575 | 0.23 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr14_+_62793175 | 0.23 |
ENSMUST00000039064.8
|
Fam124a
|
family with sequence similarity 124, member A |
| chr11_+_115921129 | 0.22 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr11_+_73068063 | 0.22 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr7_-_119744509 | 0.22 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chrX_+_165021897 | 0.22 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr9_-_40442669 | 0.21 |
ENSMUST00000119373.9
|
Gramd1b
|
GRAM domain containing 1B |
| chr7_+_44498640 | 0.20 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr12_+_81906738 | 0.20 |
ENSMUST00000221721.2
ENSMUST00000021567.6 |
Pcnx
|
pecanex homolog |
| chr7_+_44498415 | 0.20 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr5_+_33261563 | 0.20 |
ENSMUST00000011178.5
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr13_-_96678987 | 0.20 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr11_-_86561980 | 0.19 |
ENSMUST00000143991.3
|
Vmp1
|
vacuole membrane protein 1 |
| chr2_+_74566740 | 0.19 |
ENSMUST00000111982.8
|
Hoxd3
|
homeobox D3 |
| chr11_+_73067909 | 0.19 |
ENSMUST00000040687.12
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr14_+_53088747 | 0.19 |
ENSMUST00000103588.4
|
Trav13d-1
|
T cell receptor alpha variable 13D-1 |
| chr1_+_132973724 | 0.18 |
ENSMUST00000077730.7
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr13_+_19362068 | 0.18 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr5_-_142495408 | 0.18 |
ENSMUST00000110784.8
|
Radil
|
Ras association and DIL domains |
| chr14_-_34225281 | 0.18 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr7_+_138448061 | 0.17 |
ENSMUST00000041097.13
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr3_+_115801106 | 0.17 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr9_-_57342242 | 0.17 |
ENSMUST00000215961.2
|
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr3_-_115800989 | 0.16 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr18_+_90528308 | 0.16 |
ENSMUST00000235634.2
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr19_+_10366753 | 0.16 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr2_-_180798785 | 0.16 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr9_+_44990502 | 0.15 |
ENSMUST00000216426.2
|
Jaml
|
junction adhesion molecule like |
| chr6_+_57183497 | 0.15 |
ENSMUST00000227298.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr12_-_72711509 | 0.15 |
ENSMUST00000221750.2
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr5_+_138083345 | 0.15 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr13_+_96679110 | 0.15 |
ENSMUST00000179226.8
|
Cert1
|
ceramide transporter 1 |
| chr15_-_79326311 | 0.14 |
ENSMUST00000230942.2
|
Csnk1e
|
casein kinase 1, epsilon |
| chr3_-_108107729 | 0.14 |
ENSMUST00000106654.2
|
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr10_-_108535970 | 0.14 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr12_-_72711533 | 0.13 |
ENSMUST00000021512.11
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr2_-_157413189 | 0.13 |
ENSMUST00000173378.8
|
Blcap
|
bladder cancer associated protein |
| chr4_-_43700807 | 0.13 |
ENSMUST00000055545.5
|
Olfr70
|
olfactory receptor 70 |
| chrX_+_106192510 | 0.12 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr13_-_95359955 | 0.12 |
ENSMUST00000159608.8
|
Pde8b
|
phosphodiesterase 8B |
| chr9_-_35030479 | 0.12 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chrX_+_160080763 | 0.12 |
ENSMUST00000207353.2
|
Gm15262
|
predicted gene 15262 |
| chr19_+_10366450 | 0.11 |
ENSMUST00000073899.6
|
Syt7
|
synaptotagmin VII |
| chr3_+_107198618 | 0.11 |
ENSMUST00000106723.2
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chr3_+_107198528 | 0.10 |
ENSMUST00000029502.14
|
Slc16a4
|
solute carrier family 16 (monocarboxylic acid transporters), member 4 |
| chrX_-_156351979 | 0.10 |
ENSMUST00000065806.5
|
Yy2
|
Yy2 transcription factor |
| chrX_+_94942639 | 0.09 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr8_-_110419867 | 0.09 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr4_+_138606671 | 0.09 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr15_-_99829674 | 0.09 |
ENSMUST00000062631.15
|
Fam186a
|
family with sequence similarity 186, member A |
| chr9_-_121620150 | 0.09 |
ENSMUST00000215910.2
ENSMUST00000215477.2 ENSMUST00000163981.3 |
Hhatl
|
hedgehog acyltransferase-like |
| chr15_-_89012838 | 0.08 |
ENSMUST00000082197.12
|
Hdac10
|
histone deacetylase 10 |
| chr14_+_66378382 | 0.08 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr9_+_44990447 | 0.08 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chr17_+_35235552 | 0.08 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr11_-_48792973 | 0.07 |
ENSMUST00000109210.8
|
Gm5431
|
predicted gene 5431 |
| chr7_-_44497950 | 0.07 |
ENSMUST00000208890.2
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr15_+_102052797 | 0.06 |
ENSMUST00000023807.7
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr8_-_96615138 | 0.06 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr14_+_53878158 | 0.05 |
ENSMUST00000179267.4
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr14_+_53836282 | 0.05 |
ENSMUST00000103655.3
|
Trav4-3
|
T cell receptor alpha variable 4-3 |
| chr2_-_89855921 | 0.05 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr16_+_34510918 | 0.05 |
ENSMUST00000023532.7
|
Ccdc14
|
coiled-coil domain containing 14 |
| chrX_-_166047289 | 0.04 |
ENSMUST00000133722.2
|
Tlr8
|
toll-like receptor 8 |
| chr15_+_92294762 | 0.04 |
ENSMUST00000169942.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr5_+_89675288 | 0.04 |
ENSMUST00000048557.3
|
Npffr2
|
neuropeptide FF receptor 2 |
| chr2_-_157179344 | 0.04 |
ENSMUST00000109536.8
|
Ghrh
|
growth hormone releasing hormone |
| chr10_-_77618376 | 0.04 |
ENSMUST00000167358.2
|
Gm3250
|
predicted gene 3250 |
| chr11_-_16952929 | 0.03 |
ENSMUST00000156101.2
|
Plek
|
pleckstrin |
| chr1_-_162687488 | 0.03 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_-_171885386 | 0.03 |
ENSMUST00000087950.4
|
Cbln4
|
cerebellin 4 precursor protein |
| chr11_-_48793009 | 0.01 |
ENSMUST00000109212.3
|
Gm5431
|
predicted gene 5431 |
| chrX_-_153999333 | 0.01 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr7_-_44498305 | 0.01 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr13_+_68011442 | 0.01 |
ENSMUST00000078471.7
|
BC048507
|
cDNA sequence BC048507 |
| chr11_-_102076028 | 0.01 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chrX_-_72380446 | 0.00 |
ENSMUST00000207943.2
|
Gm45015
|
predicted gene 45015 |
| chr7_-_134100659 | 0.00 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chrX_-_153999440 | 0.00 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr14_+_53878403 | 0.00 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.7 | 13.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 6.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 1.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 0.6 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 1.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 3.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.5 | GO:0060618 | nipple development(GO:0060618) mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 7.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.7 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 2.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 3.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.3 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 2.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0060112 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 1.2 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.3 | GO:1904446 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0048372 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.4 | GO:0045900 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 1.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.5 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.0 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 3.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.5 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 2.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 3.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 2.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 2.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 6.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 7.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 0.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 0.9 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.3 | 3.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 1.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 2.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 3.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 11.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |