Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
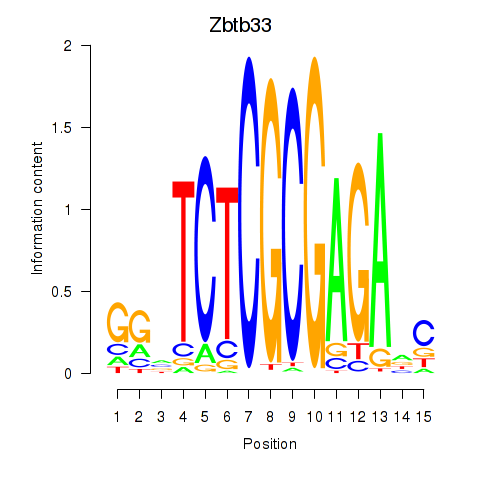
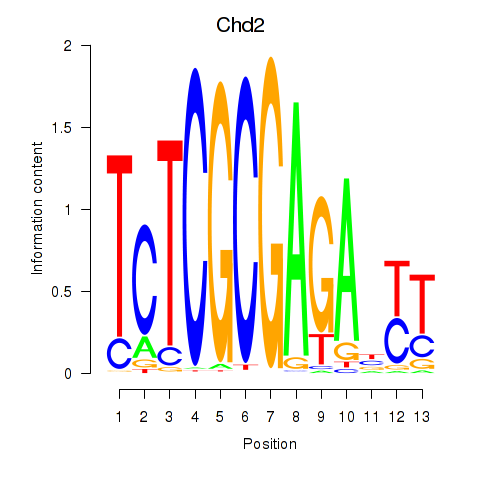
Results for Zbtb33_Chd2
Z-value: 3.40
Transcription factors associated with Zbtb33_Chd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb33
|
ENSMUSG00000048047.4 | zinc finger and BTB domain containing 33 |
|
Chd2
|
ENSMUSG00000078671.12 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Chd2 | mm39_v1_chr7_-_73191484_73191503 | 0.81 | 1.8e-09 | Click! |
| Zbtb33 | mm39_v1_chrX_+_37278636_37278722 | 0.69 | 3.1e-06 | Click! |
Activity profile of Zbtb33_Chd2 motif
Sorted Z-values of Zbtb33_Chd2 motif
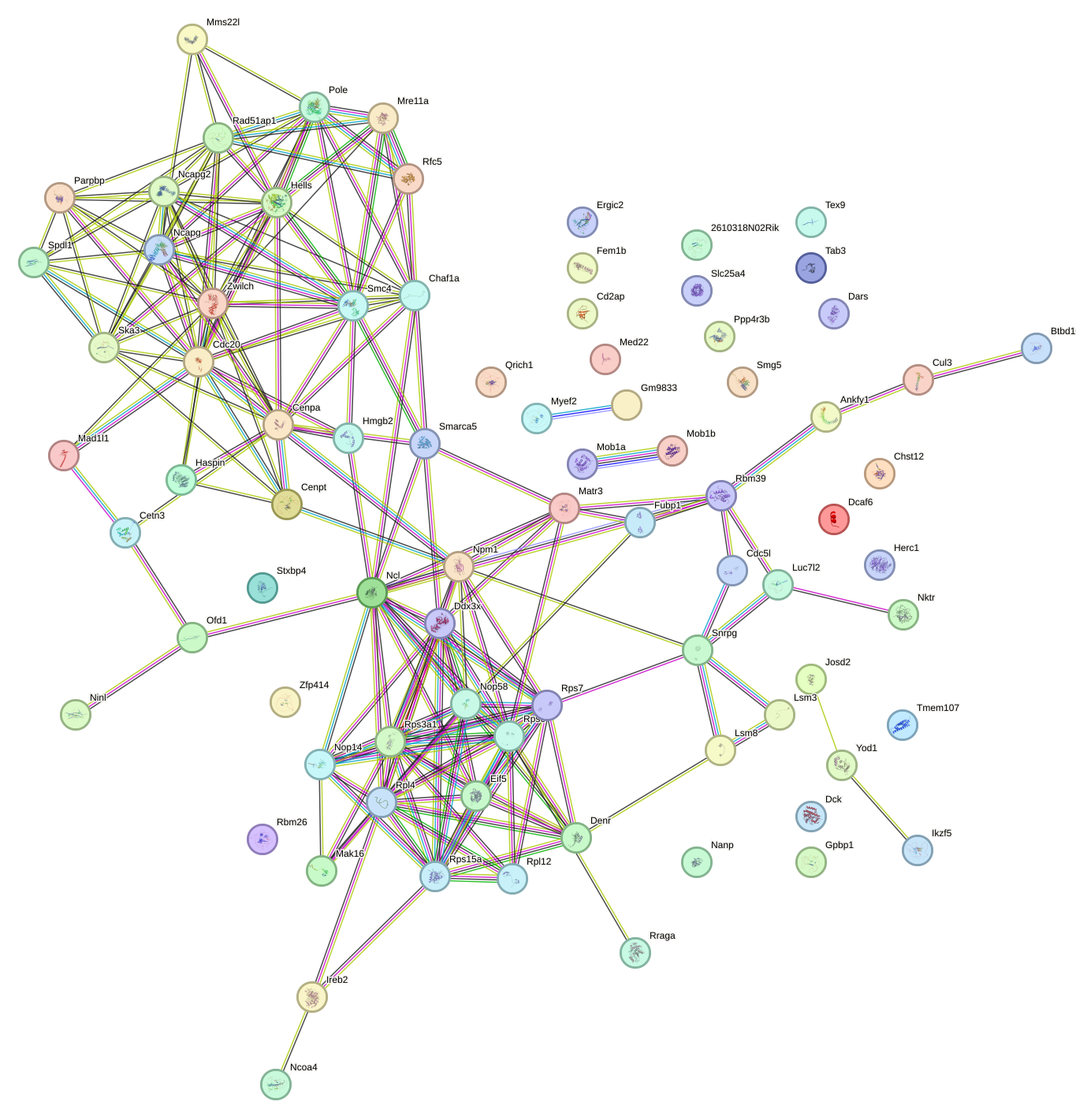
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb33_Chd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 2.9 | 11.6 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 2.7 | 8.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 2.5 | 15.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 2.2 | 8.9 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 2.2 | 8.7 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 2.1 | 10.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 2.1 | 6.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 2.0 | 6.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 2.0 | 6.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.0 | 6.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 2.0 | 7.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.8 | 18.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.8 | 21.5 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 1.8 | 5.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.8 | 5.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.7 | 10.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.7 | 6.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 1.6 | 8.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 1.6 | 12.6 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 1.5 | 6.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.5 | 4.6 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 1.5 | 5.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.5 | 11.6 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.4 | 23.1 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 1.4 | 10.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 1.4 | 6.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.3 | 7.7 | GO:0032439 | endosome localization(GO:0032439) |
| 1.2 | 13.5 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 1.2 | 12.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.1 | 7.5 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 1.0 | 2.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 1.0 | 3.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.0 | 10.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.9 | 2.8 | GO:0048822 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.9 | 5.6 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.9 | 11.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.9 | 3.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.9 | 14.5 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.8 | 5.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.8 | 4.2 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.8 | 17.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.8 | 2.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.8 | 2.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.7 | 21.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.7 | 2.8 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.7 | 2.0 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 5.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 1.9 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.6 | 1.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.6 | 5.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.6 | 1.8 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.6 | 5.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.6 | 6.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 11.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.5 | 4.4 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.5 | 18.0 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.5 | 4.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 2.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.5 | 4.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.5 | 1.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.5 | 10.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.5 | 15.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.5 | 19.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.5 | 2.7 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.4 | 4.5 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 1.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 3.5 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.4 | 3.0 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 5.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.4 | 1.7 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.4 | 3.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.4 | 0.4 | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.4 | 5.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.4 | 8.9 | GO:0072401 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.4 | 1.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 | 17.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.4 | 2.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 1.5 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 | 2.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 10.1 | GO:0045005 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.4 | 2.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 6.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 1.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.4 | 3.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 14.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 3.6 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 3.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 2.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.3 | 19.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 3.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.3 | 3.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 0.8 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 | 8.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 1.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.3 | 2.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 6.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 5.0 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.3 | 8.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 3.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 0.7 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.2 | 1.9 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 9.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 2.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 3.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 10.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 3.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 1.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 1.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 4.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 4.8 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.2 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.8 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.2 | 9.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 2.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 0.3 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.2 | 1.0 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 3.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 3.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 4.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 3.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 4.9 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 2.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 2.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 2.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 3.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.3 | GO:0022028 | succinyl-CoA metabolic process(GO:0006104) tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 18.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 2.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 1.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 0.8 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 5.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.8 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 4.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 3.6 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 0.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 6.8 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 10.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 3.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 7.9 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 24.8 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 18.2 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.1 | 0.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.5 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 12.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 0.3 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.3 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 1.0 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 9.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 3.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 3.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 1.0 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 1.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 6.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.8 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 1.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 7.1 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.4 | 10.3 | GO:0070992 | translation initiation complex(GO:0070992) |
| 3.4 | 40.3 | GO:0000796 | condensin complex(GO:0000796) |
| 2.4 | 9.5 | GO:0035101 | FACT complex(GO:0035101) |
| 2.0 | 6.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.7 | 15.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 1.7 | 10.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.6 | 6.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.5 | 9.2 | GO:0001652 | granular component(GO:0001652) |
| 1.5 | 22.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.4 | 6.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.3 | 18.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 1.3 | 5.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.2 | 10.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.2 | 8.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.2 | 5.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.1 | 3.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.0 | 6.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.0 | 10.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 1.0 | 5.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.0 | 59.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.8 | 7.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 2.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.8 | 4.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.7 | 11.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.7 | 4.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.7 | 15.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.7 | 2.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.7 | 2.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.6 | 11.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.6 | 1.9 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.6 | 6.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 4.9 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.6 | 4.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.6 | 4.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 8.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 13.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 39.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 8.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 6.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.5 | 7.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 20.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.5 | 8.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.5 | 6.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 5.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 5.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 5.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 10.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 2.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 2.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) telomeric heterochromatin(GO:0031933) |
| 0.3 | 8.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 2.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 3.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 4.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 1.9 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 2.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 9.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 5.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 6.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 3.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 2.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.7 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 5.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 4.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 4.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 12.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 11.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 7.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.4 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 1.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 3.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 7.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.2 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 4.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 3.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 5.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 6.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 3.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 2.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 5.1 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 26.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 7.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 3.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 7.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 10.8 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 37.3 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 2.4 | 11.9 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 2.2 | 15.5 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 2.1 | 6.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 2.0 | 6.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.9 | 7.8 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 1.8 | 12.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.7 | 11.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.7 | 5.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.5 | 8.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.4 | 11.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.4 | 11.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.3 | 5.3 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 1.1 | 14.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.1 | 11.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.0 | 11.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.9 | 2.8 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.8 | 3.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.8 | 5.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.7 | 12.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 13.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.7 | 4.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.7 | 8.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 7.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 2.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.6 | 6.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.6 | 6.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.6 | 2.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.6 | 3.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.6 | 2.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.6 | 5.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 4.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 5.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.5 | 5.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 5.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 6.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.5 | 3.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 22.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.5 | 3.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.5 | 6.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 1.3 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.4 | 1.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 2.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.4 | 18.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.4 | 4.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 2.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.3 | 14.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.3 | 3.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 15.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 1.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 8.2 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.3 | 4.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 5.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 1.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.3 | 9.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 1.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 5.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 2.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 6.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 21.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 8.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 16.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 10.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 1.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 4.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 1.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 11.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 20.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 31.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 3.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 3.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 4.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 5.2 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 6.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.5 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 8.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 9.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 16.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 14.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 4.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 5.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 14.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 5.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 5.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 8.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0032795 | thioesterase binding(GO:0031996) heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.4 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 72.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 14.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 15.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 8.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 11.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 10.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 5.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 10.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 10.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 7.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 7.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 7.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 14.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.0 | 22.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.7 | 83.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.6 | 9.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.6 | 19.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.6 | 6.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.5 | 10.8 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.5 | 9.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.5 | 11.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 14.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 5.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 12.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 11.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.3 | 34.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 3.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 3.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 2.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.3 | 6.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 5.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 5.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 3.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 11.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 16.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 4.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 10.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.3 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 4.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |