Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
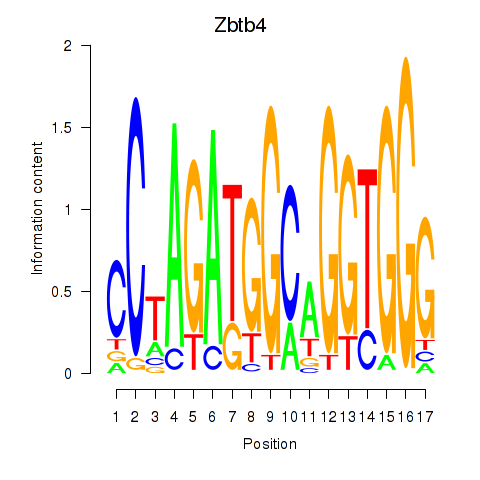
Results for Zbtb4
Z-value: 1.24
Transcription factors associated with Zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb4
|
ENSMUSG00000018750.15 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | mm39_v1_chr11_+_69657275_69657385 | 0.43 | 8.4e-03 | Click! |
Activity profile of Zbtb4 motif
Sorted Z-values of Zbtb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90603013 | 18.11 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr7_-_140596811 | 9.79 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chrX_-_92875712 | 6.65 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr16_-_22946441 | 6.61 |
ENSMUST00000133847.9
ENSMUST00000115338.8 ENSMUST00000023598.15 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr8_+_84682136 | 6.51 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr4_-_156340276 | 6.38 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr14_+_30856687 | 5.84 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr15_+_73594965 | 5.66 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr15_+_73595012 | 5.55 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr9_+_69361348 | 4.84 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr7_-_125968653 | 4.67 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr2_+_29759495 | 4.56 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr4_-_156340713 | 4.53 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr6_-_57802131 | 4.31 |
ENSMUST00000204878.3
ENSMUST00000145608.7 ENSMUST00000203212.3 ENSMUST00000114297.5 |
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_-_171061838 | 4.08 |
ENSMUST00000193973.2
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr11_-_69786324 | 4.04 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_+_39472981 | 3.80 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr6_-_126916487 | 3.70 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr17_-_48716756 | 3.56 |
ENSMUST00000160319.8
ENSMUST00000159535.2 ENSMUST00000078800.13 ENSMUST00000046719.14 ENSMUST00000162460.8 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr17_-_26161797 | 3.54 |
ENSMUST00000208043.2
ENSMUST00000148382.2 ENSMUST00000145745.3 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_96078340 | 3.52 |
ENSMUST00000034982.16
ENSMUST00000188008.7 ENSMUST00000188750.7 ENSMUST00000185644.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr7_+_28682253 | 3.46 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr1_+_39406979 | 3.37 |
ENSMUST00000178079.8
ENSMUST00000179954.8 |
Rpl31
|
ribosomal protein L31 |
| chr11_-_101998648 | 3.34 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr14_-_30348153 | 3.32 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chr4_+_130253925 | 3.07 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr4_+_6365694 | 3.02 |
ENSMUST00000175769.8
ENSMUST00000140830.8 ENSMUST00000108374.8 |
Sdcbp
|
syndecan binding protein |
| chrX_-_47551990 | 3.01 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr17_+_29020064 | 2.97 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr1_+_39406954 | 2.95 |
ENSMUST00000194746.6
|
Rpl31
|
ribosomal protein L31 |
| chr11_-_3454766 | 2.92 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr1_-_171061902 | 2.85 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr9_+_106080307 | 2.83 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chrX_-_132799144 | 2.76 |
ENSMUST00000087557.12
|
Tspan6
|
tetraspanin 6 |
| chr9_+_118881838 | 2.72 |
ENSMUST00000051386.13
ENSMUST00000074734.13 |
Vill
|
villin-like |
| chr4_+_6365650 | 2.69 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr8_-_78451055 | 2.60 |
ENSMUST00000034029.8
|
Ednra
|
endothelin receptor type A |
| chr5_+_110434172 | 2.59 |
ENSMUST00000007296.12
ENSMUST00000112482.2 |
Pole
|
polymerase (DNA directed), epsilon |
| chr1_+_171216480 | 2.58 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr7_-_24705320 | 2.56 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr1_+_39407183 | 2.50 |
ENSMUST00000195123.6
|
Rpl31
|
ribosomal protein L31 |
| chr2_-_84500951 | 2.44 |
ENSMUST00000189988.3
ENSMUST00000189636.8 ENSMUST00000102646.4 ENSMUST00000102647.11 ENSMUST00000117299.10 |
Selenoh
|
selenoprotein H |
| chr8_-_65146079 | 2.42 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr9_+_96078299 | 2.42 |
ENSMUST00000165120.9
|
Tfdp2
|
transcription factor Dp 2 |
| chr10_+_80765900 | 2.39 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr2_+_30306116 | 2.37 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr7_-_99002204 | 2.36 |
ENSMUST00000208292.2
ENSMUST00000207989.2 ENSMUST00000208749.2 ENSMUST00000169437.9 ENSMUST00000208119.2 ENSMUST00000207849.2 |
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr1_-_180641430 | 2.34 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr9_+_106158549 | 2.33 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr19_+_45035942 | 2.32 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chrX_-_132799041 | 2.31 |
ENSMUST00000176718.8
ENSMUST00000176641.2 |
Tspan6
|
tetraspanin 6 |
| chr11_-_115968576 | 2.31 |
ENSMUST00000106450.8
|
Unc13d
|
unc-13 homolog D |
| chr15_+_76235503 | 2.26 |
ENSMUST00000023212.15
ENSMUST00000160172.8 |
Maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr2_+_30306045 | 2.18 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr2_-_30305472 | 2.15 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr9_+_106158212 | 2.14 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr7_-_34088886 | 2.14 |
ENSMUST00000155256.8
|
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr11_+_75542902 | 2.10 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr7_+_25872836 | 2.08 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr19_+_45036037 | 2.08 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr2_+_124452194 | 2.05 |
ENSMUST00000051419.15
ENSMUST00000076335.12 ENSMUST00000078621.12 ENSMUST00000077847.12 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr13_-_55661240 | 2.05 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr9_-_37166699 | 2.04 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr1_-_93270430 | 2.02 |
ENSMUST00000027493.4
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr11_+_77384234 | 1.99 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr3_-_83749036 | 1.95 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr3_-_152045986 | 1.94 |
ENSMUST00000199397.2
ENSMUST00000199334.5 ENSMUST00000068243.11 ENSMUST00000073089.13 |
Miga1
|
mitoguardin 1 |
| chr7_-_35096133 | 1.94 |
ENSMUST00000154597.2
ENSMUST00000032704.12 |
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chrX_+_163156359 | 1.93 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr11_+_121593219 | 1.92 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr10_-_35587888 | 1.91 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr6_-_128332789 | 1.88 |
ENSMUST00000001562.9
|
Tulp3
|
tubby-like protein 3 |
| chr9_-_107512511 | 1.86 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr14_+_63673843 | 1.78 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr11_-_69470139 | 1.78 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr9_-_107512566 | 1.77 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr16_+_22713593 | 1.76 |
ENSMUST00000232674.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr1_-_152262425 | 1.76 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr2_-_170269748 | 1.73 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr2_+_25313240 | 1.70 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr9_+_108820846 | 1.70 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr14_+_121148625 | 1.70 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr11_-_115968373 | 1.66 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr4_+_119052693 | 1.64 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr17_-_29483075 | 1.63 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr7_-_27252543 | 1.61 |
ENSMUST00000127240.8
ENSMUST00000117095.8 ENSMUST00000117611.8 |
Pld3
|
phospholipase D family, member 3 |
| chr19_-_11582207 | 1.60 |
ENSMUST00000025582.11
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr11_+_75542328 | 1.57 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr18_+_42644552 | 1.56 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr2_-_144112444 | 1.56 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr8_+_95807814 | 1.55 |
ENSMUST00000034239.9
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chr1_-_36978602 | 1.54 |
ENSMUST00000027290.12
|
Tmem131
|
transmembrane protein 131 |
| chr2_+_124452493 | 1.49 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr19_+_45036220 | 1.49 |
ENSMUST00000084493.8
|
Sfxn3
|
sideroflexin 3 |
| chr11_-_69728560 | 1.47 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr5_-_123126550 | 1.46 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr7_+_27252658 | 1.46 |
ENSMUST00000067386.14
ENSMUST00000191126.7 ENSMUST00000187960.7 |
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_+_126446588 | 1.45 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr12_+_102521225 | 1.38 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr4_+_124960465 | 1.37 |
ENSMUST00000052183.7
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr6_-_49240944 | 1.36 |
ENSMUST00000204189.3
ENSMUST00000031841.9 |
Tra2a
|
transformer 2 alpha |
| chr8_+_107237483 | 1.32 |
ENSMUST00000080797.8
|
Cdh3
|
cadherin 3 |
| chr4_+_156194427 | 1.32 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr1_-_152262339 | 1.32 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr8_-_123302187 | 1.29 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chr8_+_105065980 | 1.25 |
ENSMUST00000212139.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr17_-_74601769 | 1.24 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr2_-_30305779 | 1.19 |
ENSMUST00000102855.8
ENSMUST00000028207.13 |
Crat
|
carnitine acetyltransferase |
| chr1_-_136161850 | 1.19 |
ENSMUST00000120339.8
|
Inava
|
innate immunity activator |
| chr17_+_29833760 | 1.18 |
ENSMUST00000024817.15
ENSMUST00000162588.3 |
Rnf8
|
ring finger protein 8 |
| chr11_-_3477916 | 1.16 |
ENSMUST00000020718.10
|
Smtn
|
smoothelin |
| chr14_+_30723340 | 1.16 |
ENSMUST00000168584.9
ENSMUST00000226378.2 |
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr3_-_69506293 | 1.15 |
ENSMUST00000061826.3
|
B3galnt1
|
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
| chr17_-_74601828 | 1.14 |
ENSMUST00000233514.3
|
Memo1
|
mediator of cell motility 1 |
| chr3_-_144425819 | 1.12 |
ENSMUST00000199531.5
ENSMUST00000199854.5 |
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr14_+_65612788 | 1.11 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr4_-_132649798 | 1.10 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr9_+_75221415 | 1.09 |
ENSMUST00000215875.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr9_-_21199232 | 1.07 |
ENSMUST00000184326.8
ENSMUST00000038671.10 |
Kri1
|
KRI1 homolog |
| chr11_-_115967873 | 1.06 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr9_-_106035308 | 1.05 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr9_+_69902697 | 1.05 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr4_+_119052548 | 1.05 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr14_+_30723371 | 1.03 |
ENSMUST00000022476.9
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr7_+_19115929 | 1.02 |
ENSMUST00000062831.16
ENSMUST00000108461.8 ENSMUST00000108460.8 |
Ercc2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr2_+_179899166 | 1.02 |
ENSMUST00000059080.7
|
Rps21
|
ribosomal protein S21 |
| chr17_-_37269425 | 0.99 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr7_-_45045097 | 0.98 |
ENSMUST00000211121.2
ENSMUST00000074575.11 |
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr13_+_35925296 | 0.98 |
ENSMUST00000163595.3
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr7_-_34089109 | 0.95 |
ENSMUST00000085585.12
|
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr13_+_69957960 | 0.94 |
ENSMUST00000022089.10
|
Med10
|
mediator complex subunit 10 |
| chr7_-_141925947 | 0.93 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr3_-_135373560 | 0.92 |
ENSMUST00000164430.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr10_-_60055082 | 0.90 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr9_-_106035332 | 0.90 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr1_-_165064455 | 0.87 |
ENSMUST00000043235.8
|
Tiprl
|
TIP41, TOR signalling pathway regulator-like (S. cerevisiae) |
| chr15_-_58695379 | 0.83 |
ENSMUST00000072113.6
|
Tmem65
|
transmembrane protein 65 |
| chr13_+_85337552 | 0.82 |
ENSMUST00000165077.8
ENSMUST00000164127.8 ENSMUST00000163600.8 |
Ccnh
|
cyclin H |
| chr16_+_32250043 | 0.81 |
ENSMUST00000115140.2
ENSMUST00000104893.10 |
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr9_+_118881926 | 0.80 |
ENSMUST00000131647.2
|
Vill
|
villin-like |
| chr3_+_127583450 | 0.80 |
ENSMUST00000054483.14
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr7_-_35346553 | 0.80 |
ENSMUST00000120714.2
|
Pdcd5
|
programmed cell death 5 |
| chr5_+_145020640 | 0.79 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr14_-_30723292 | 0.78 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr12_+_111505216 | 0.78 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr5_+_21087107 | 0.77 |
ENSMUST00000115259.3
|
Tmem60
|
transmembrane protein 60 |
| chr5_+_145020910 | 0.77 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr9_-_8042785 | 0.76 |
ENSMUST00000215478.2
ENSMUST00000065291.2 |
Cfap300
|
cilia and flagella associated protein 300 |
| chr19_-_40600619 | 0.76 |
ENSMUST00000132452.2
ENSMUST00000135795.8 ENSMUST00000025981.15 |
Tctn3
|
tectonic family member 3 |
| chr9_+_57924679 | 0.76 |
ENSMUST00000188539.2
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr18_-_77802145 | 0.75 |
ENSMUST00000237203.2
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr2_-_71198091 | 0.75 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr10_+_4216353 | 0.75 |
ENSMUST00000045730.7
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr18_+_11790409 | 0.75 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr7_-_144024451 | 0.74 |
ENSMUST00000033407.13
|
Cttn
|
cortactin |
| chr17_+_32671689 | 0.74 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr7_+_16472335 | 0.73 |
ENSMUST00000086112.8
ENSMUST00000205607.2 |
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr4_-_6454097 | 0.73 |
ENSMUST00000124344.2
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr4_-_130253703 | 0.71 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr3_+_127583550 | 0.69 |
ENSMUST00000163775.6
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr8_+_106785434 | 0.69 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr7_+_15909283 | 0.68 |
ENSMUST00000002495.18
|
Meis3
|
Meis homeobox 3 |
| chr5_+_136112765 | 0.68 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr2_-_134486039 | 0.67 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr3_-_86828140 | 0.66 |
ENSMUST00000029719.14
|
Dclk2
|
doublecortin-like kinase 2 |
| chr7_-_35346889 | 0.64 |
ENSMUST00000118501.8
|
Pdcd5
|
programmed cell death 5 |
| chr14_-_54651442 | 0.64 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr7_+_140547941 | 0.63 |
ENSMUST00000106040.8
ENSMUST00000026564.9 |
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr18_-_77801624 | 0.63 |
ENSMUST00000074653.6
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr17_-_37269330 | 0.62 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr4_-_88595161 | 0.62 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr5_-_77099406 | 0.62 |
ENSMUST00000140076.2
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr12_+_111505253 | 0.60 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr14_+_75253453 | 0.60 |
ENSMUST00000036072.8
|
Rubcnl
|
RUN and cysteine rich domain containing beclin 1 interacting protein like |
| chr1_+_183078851 | 0.60 |
ENSMUST00000193625.2
|
Aida
|
axin interactor, dorsalization associated |
| chr7_-_103778992 | 0.59 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr19_-_46384158 | 0.58 |
ENSMUST00000237742.2
ENSMUST00000040270.6 |
Actr1a
|
ARP1 actin-related protein 1A, centractin alpha |
| chr9_-_106764627 | 0.58 |
ENSMUST00000055843.9
|
Rbm15b
|
RNA binding motif protein 15B |
| chr6_+_70549568 | 0.58 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr14_-_30723549 | 0.58 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr3_-_129854477 | 0.57 |
ENSMUST00000001079.15
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr17_+_87944110 | 0.57 |
ENSMUST00000234623.2
|
Epcam
|
epithelial cell adhesion molecule |
| chr8_-_120828211 | 0.56 |
ENSMUST00000034280.9
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr13_+_55357585 | 0.55 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr6_-_38853097 | 0.54 |
ENSMUST00000161779.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr17_+_6926452 | 0.54 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr2_-_84652890 | 0.53 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr6_+_68196928 | 0.52 |
ENSMUST00000103318.6
ENSMUST00000103319.3 |
Igkv2-112
|
immunoglobulin kappa variable 2-112 |
| chr15_-_89033761 | 0.52 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr5_+_117457126 | 0.52 |
ENSMUST00000111967.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr7_-_79585006 | 0.51 |
ENSMUST00000048731.6
|
Arpin
|
actin-related protein 2/3 complex inhibitor |
| chr15_-_43146092 | 0.50 |
ENSMUST00000022960.4
|
Eif3e
|
eukaryotic translation initiation factor 3, subunit E |
| chr19_-_5438685 | 0.50 |
ENSMUST00000044207.5
|
Sart1
|
squamous cell carcinoma antigen recognized by T cells 1 |
| chr11_-_22810467 | 0.49 |
ENSMUST00000055549.4
|
B3gnt2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr17_+_8502594 | 0.48 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr16_-_43800109 | 0.47 |
ENSMUST00000231700.2
|
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr16_+_20511991 | 0.47 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr6_-_38852857 | 0.46 |
ENSMUST00000162359.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr2_-_134485936 | 0.45 |
ENSMUST00000110120.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr3_+_65435825 | 0.44 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.7 | 6.9 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 1.7 | 6.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.5 | 4.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.0 | 2.0 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 1.0 | 5.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.9 | 2.6 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.8 | 3.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.8 | 2.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.8 | 4.8 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.7 | 6.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.6 | 1.9 | GO:0042495 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.6 | 1.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.6 | 5.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.6 | 6.8 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.6 | 6.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 10.9 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.5 | 1.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.5 | 1.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.5 | 2.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.4 | 3.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 2.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.4 | 1.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.4 | GO:2000705 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 2.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 1.3 | GO:0046084 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.3 | 1.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 5.1 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.3 | 0.9 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.3 | 3.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 2.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.3 | 2.8 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 0.8 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.3 | 1.3 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.3 | 4.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 1.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 2.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 3.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 2.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 3.3 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.2 | 1.8 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 3.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 2.6 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.2 | 1.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 2.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.6 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.2 | 1.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 3.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 3.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 2.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 1.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 8.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.9 | GO:0070535 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 1.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.0 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 3.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.7 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 0.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 3.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 1.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 3.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) response to stilbenoid(GO:0035634) |
| 0.1 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 3.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 5.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 1.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 2.2 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.0 | 0.9 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.0 | GO:1905214 | regulation of mRNA binding(GO:1902415) regulation of RNA binding(GO:1905214) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 1.0 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.0 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 1.6 | 4.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.1 | 6.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.7 | 3.7 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.7 | 3.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.6 | 1.9 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.6 | 2.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.5 | 2.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 5.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 3.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 0.8 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.3 | 2.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 2.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 3.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.6 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.2 | 1.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 4.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 9.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 3.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 4.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 7.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 4.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 10.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 4.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 8.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 2.3 | 18.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.9 | 5.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.7 | 6.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 6.9 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 1.1 | 4.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.0 | 2.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.9 | 2.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 3.3 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.7 | 2.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.6 | 1.9 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.6 | 6.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 4.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.5 | 3.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.5 | 1.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.5 | 2.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.4 | 1.3 | GO:0002055 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.3 | 1.8 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.3 | 2.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 1.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 2.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 3.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 5.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 4.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.8 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.2 | 1.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.9 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 1.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 3.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 3.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.4 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 1.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 3.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 4.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 11.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 3.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 9.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 1.1 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 7.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 8.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 4.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 6.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 19.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 8.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 11.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 3.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 9.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 6.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 7.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 7.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 6.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 9.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.4 | 6.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 3.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 9.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 4.4 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 4.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 5.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |