Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zbtb7a
Z-value: 1.69
Transcription factors associated with Zbtb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7a
|
ENSMUSG00000035011.16 | zinc finger and BTB domain containing 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7a | mm39_v1_chr10_+_80972089_80972105 | 0.76 | 8.7e-08 | Click! |
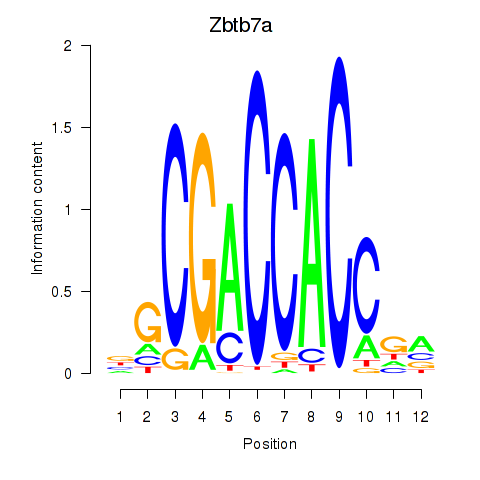
Activity profile of Zbtb7a motif
Sorted Z-values of Zbtb7a motif
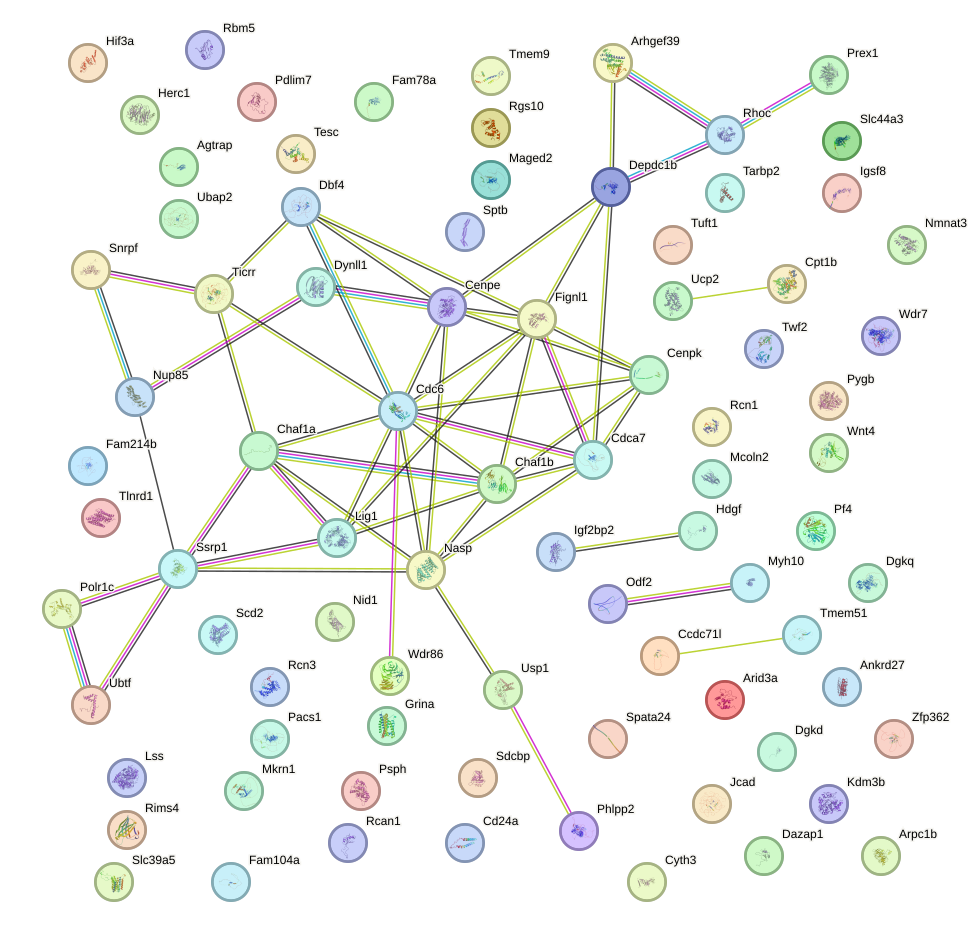
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 6.7 | 33.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 5.3 | 16.0 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 2.4 | 28.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 1.7 | 13.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.6 | 8.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.5 | 6.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.4 | 10.9 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.3 | 8.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.3 | 3.9 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.3 | 13.9 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.2 | 4.7 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 1.1 | 3.4 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 1.0 | 2.9 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.9 | 18.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.9 | 9.5 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.8 | 5.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.8 | 2.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.8 | 3.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.8 | 3.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 2.9 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.7 | 5.7 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.6 | 1.9 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.6 | 4.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 1.8 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.6 | 7.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 4.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.6 | 1.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.6 | 2.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.6 | 6.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.6 | 2.8 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.5 | 7.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.5 | 1.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.5 | 8.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 2.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 2.9 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.5 | 4.8 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.5 | 1.4 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.4 | 2.7 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.4 | 2.7 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.4 | 2.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.4 | 4.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.4 | 2.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 1.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 5.8 | GO:0009650 | UV protection(GO:0009650) |
| 0.4 | 2.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.4 | 1.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.3 | 1.4 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.3 | 3.4 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.3 | 1.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 1.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.3 | 1.0 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.3 | 11.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.3 | 5.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 2.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.3 | 2.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 0.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 6.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.3 | 1.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 7.1 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.3 | 1.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 1.0 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 3.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 2.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 2.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 2.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 1.2 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.2 | 1.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 10.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.2 | 7.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 2.2 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.2 | 1.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 1.5 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 2.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 2.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 1.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 1.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.2 | 2.4 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 0.6 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 1.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 6.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 0.8 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.2 | 8.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 2.4 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.2 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 7.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.0 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 3.0 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.8 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.4 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.4 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 4.1 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 1.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.6 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 3.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 3.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 1.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 3.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 2.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 2.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.6 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 5.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.7 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 8.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 4.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.6 | GO:1904778 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.2 | GO:0097477 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 3.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 1.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 9.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 1.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 0.5 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 3.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 0.2 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.0 | 2.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 4.9 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.2 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 3.5 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 8.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.1 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 1.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 3.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.0 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 1.7 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 2.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 2.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.5 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 7.8 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 7.9 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 1.2 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 1.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 8.9 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.8 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 1.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 2.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 4.0 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.6 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 2.7 | 10.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.4 | 33.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.1 | 16.7 | GO:0008091 | spectrin(GO:0008091) |
| 1.4 | 5.5 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 1.1 | 3.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.9 | 6.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.9 | 5.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.8 | 2.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.7 | 3.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.7 | 2.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.6 | 16.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 2.3 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.6 | 2.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 2.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.5 | 3.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 1.5 | GO:0090537 | CERF complex(GO:0090537) |
| 0.5 | 2.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.4 | 2.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 8.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 3.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 2.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 1.8 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 4.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 1.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 1.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 1.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 7.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 4.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 6.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 3.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.7 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.2 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 2.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 2.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 3.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 8.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 0.6 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 2.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 3.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 0.5 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 10.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 8.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 4.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 3.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 6.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 9.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 3.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.2 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 16.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 1.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 5.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 6.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 8.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 4.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 3.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 5.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 33.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.9 | 17.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.9 | 28.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.7 | 5.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.5 | 6.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.3 | 8.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.3 | 7.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.2 | 3.6 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 1.1 | 5.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.0 | 4.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.0 | 9.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 4.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.8 | 2.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.7 | 2.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.7 | 2.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.7 | 4.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.6 | 3.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.6 | 2.8 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.6 | 12.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 1.6 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.5 | 5.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.5 | 8.4 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.5 | 3.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.3 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 2.7 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.4 | 2.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.4 | 1.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 2.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 2.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 1.4 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.4 | 1.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 2.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 15.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 2.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 2.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 8.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 3.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 2.9 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.3 | 2.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 22.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 2.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 1.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 4.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 3.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 4.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 2.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 6.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 8.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.4 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.2 | 1.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.6 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.2 | 0.8 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 1.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 4.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 2.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 3.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 2.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 3.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 8.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0047238 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 2.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 25.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.6 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 7.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.9 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 3.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 2.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 3.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 10.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 5.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 16.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 8.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 8.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.1 | 4.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 3.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 6.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 2.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.0 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 3.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.8 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 1.0 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 36.2 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 5.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 35.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 9.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 15.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 3.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 7.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 7.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 6.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 5.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 10.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 33.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 11.4 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.5 | 7.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 6.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.4 | 6.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 5.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 3.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 9.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 11.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 4.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.3 | 12.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 8.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 6.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.3 | 2.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 6.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 3.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 6.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 2.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 6.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 6.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 12.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 2.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 16.7 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 4.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 6.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 7.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 9.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 2.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 5.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 19.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.4 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 4.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 3.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |