Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
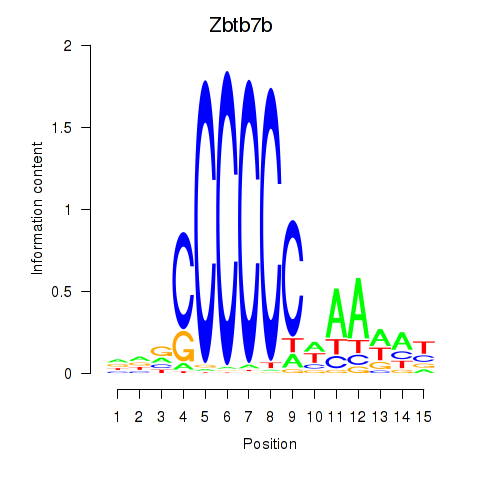
Results for Zbtb7b
Z-value: 0.57
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSMUSG00000028042.16 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | mm39_v1_chr3_-_89300936_89300992 | -0.31 | 6.8e-02 | Click! |
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_33136021 | 2.34 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr6_-_141892686 | 1.67 |
ENSMUST00000042119.6
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr6_-_141892517 | 1.56 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr7_-_126062272 | 1.43 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr3_-_107893676 | 1.32 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr7_-_28465870 | 1.25 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr7_-_140590605 | 1.09 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chrX_+_93278588 | 1.06 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chrX_+_93278526 | 1.01 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr1_+_155433858 | 0.99 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr11_-_113600346 | 0.83 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr15_-_76501041 | 0.73 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr15_-_50753792 | 0.71 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_+_139243012 | 0.65 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr7_-_34353767 | 0.64 |
ENSMUST00000206501.2
ENSMUST00000108069.8 |
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr11_-_113600838 | 0.61 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr12_-_83643964 | 0.58 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr7_+_28466160 | 0.57 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr19_-_6887361 | 0.57 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr12_-_83643883 | 0.57 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr11_-_100026754 | 0.57 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr11_-_82655132 | 0.56 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr6_+_116241146 | 0.55 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr15_-_74508197 | 0.53 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr17_-_13179589 | 0.52 |
ENSMUST00000233792.2
ENSMUST00000007005.14 |
Acat2
|
acetyl-Coenzyme A acetyltransferase 2 |
| chr10_+_84591919 | 0.52 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr11_-_115078653 | 0.51 |
ENSMUST00000103041.8
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_42420216 | 0.51 |
ENSMUST00000048630.8
ENSMUST00000238290.2 |
Crtac1
|
cartilage acidic protein 1 |
| chr19_+_36532061 | 0.49 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr1_-_170978157 | 0.48 |
ENSMUST00000155798.2
ENSMUST00000081560.5 ENSMUST00000111336.10 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr9_-_107215504 | 0.48 |
ENSMUST00000118051.2
ENSMUST00000035196.14 |
Hemk1
|
HemK methyltransferase family member 1 |
| chr19_-_6886898 | 0.47 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr19_-_6886965 | 0.47 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr8_+_84699580 | 0.45 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr17_-_35454729 | 0.45 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr5_+_16139683 | 0.44 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr15_-_76127600 | 0.43 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr11_-_120508713 | 0.43 |
ENSMUST00000106188.4
ENSMUST00000026129.16 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_+_8816663 | 0.41 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr7_-_30754223 | 0.40 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30754240 | 0.40 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr16_+_41353360 | 0.39 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chrX_-_100838004 | 0.39 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr11_-_103109247 | 0.39 |
ENSMUST00000103076.2
|
Spata32
|
spermatogenesis associated 32 |
| chrX_+_35861851 | 0.38 |
ENSMUST00000073339.7
|
Pgrmc1
|
progesterone receptor membrane component 1 |
| chr2_-_73605387 | 0.38 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr9_-_103182246 | 0.38 |
ENSMUST00000142540.2
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr11_+_50917831 | 0.38 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr7_-_4525793 | 0.37 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr7_+_55443967 | 0.37 |
ENSMUST00000206454.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr7_+_43441315 | 0.37 |
ENSMUST00000005891.7
|
Klk9
|
kallikrein related-peptidase 9 |
| chr8_-_106140106 | 0.36 |
ENSMUST00000167294.8
ENSMUST00000063071.13 |
Kctd19
|
potassium channel tetramerisation domain containing 19 |
| chr11_-_97909134 | 0.36 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr19_-_6969845 | 0.36 |
ENSMUST00000025915.13
|
Dnajc4
|
DnaJ heat shock protein family (Hsp40) member C4 |
| chr9_+_110075133 | 0.34 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr12_-_84497718 | 0.33 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr2_+_25346841 | 0.33 |
ENSMUST00000114265.9
ENSMUST00000102918.3 |
Clic3
|
chloride intracellular channel 3 |
| chr1_+_40123858 | 0.33 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr17_-_28569721 | 0.33 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr11_-_102047165 | 0.33 |
ENSMUST00000021296.7
|
Tmem101
|
transmembrane protein 101 |
| chr5_+_115568002 | 0.32 |
ENSMUST00000067168.9
|
Msi1
|
musashi RNA-binding protein 1 |
| chr11_+_70416185 | 0.32 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr1_-_74974707 | 0.32 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr15_+_92495007 | 0.32 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr5_-_132570710 | 0.32 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr13_-_112788829 | 0.31 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chr1_+_75213171 | 0.31 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr15_+_76152276 | 0.31 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr3_+_88624194 | 0.30 |
ENSMUST00000172252.2
|
Rit1
|
Ras-like without CAAX 1 |
| chr7_-_133966588 | 0.30 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr3_+_66127330 | 0.30 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr11_-_42072990 | 0.30 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr13_-_48779047 | 0.30 |
ENSMUST00000222028.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr9_-_121668527 | 0.29 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr1_+_167135933 | 0.29 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr2_+_156562989 | 0.29 |
ENSMUST00000000094.14
|
Dlgap4
|
DLG associated protein 4 |
| chr17_-_37269330 | 0.29 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr3_+_88624145 | 0.29 |
ENSMUST00000029692.15
ENSMUST00000171645.8 |
Rit1
|
Ras-like without CAAX 1 |
| chr1_+_75213114 | 0.29 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr7_-_30754792 | 0.29 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chrX_+_165021897 | 0.29 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chrX_+_165021919 | 0.28 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr3_-_95125051 | 0.28 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr11_+_108811626 | 0.28 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr10_+_34173426 | 0.28 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr4_-_98712077 | 0.27 |
ENSMUST00000097964.3
|
I0C0044D17Rik
|
RIKEN cDNA I0C0044D17 gene |
| chr1_-_25868788 | 0.27 |
ENSMUST00000151309.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr17_-_37269425 | 0.27 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr15_-_81756076 | 0.27 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr11_+_85061922 | 0.27 |
ENSMUST00000018623.4
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chr1_-_25868592 | 0.27 |
ENSMUST00000135518.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr3_+_34704554 | 0.27 |
ENSMUST00000099151.6
|
Sox2
|
SRY (sex determining region Y)-box 2 |
| chr2_-_32665596 | 0.27 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_-_83193412 | 0.27 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr2_-_32665637 | 0.27 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_+_72098363 | 0.26 |
ENSMUST00000021158.4
|
Txndc17
|
thioredoxin domain containing 17 |
| chr2_-_160155536 | 0.26 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr7_+_78432867 | 0.26 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr17_-_28569574 | 0.25 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr9_-_38899578 | 0.25 |
ENSMUST00000216823.2
ENSMUST00000214324.3 |
Olfr934
|
olfactory receptor 934 |
| chr11_+_69909659 | 0.25 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr11_-_59340739 | 0.25 |
ENSMUST00000136436.2
ENSMUST00000150297.2 ENSMUST00000010038.10 ENSMUST00000156146.8 ENSMUST00000132969.8 ENSMUST00000120940.8 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr4_-_43771009 | 0.24 |
ENSMUST00000053931.2
|
Olfr159
|
olfactory receptor 159 |
| chr7_-_19595221 | 0.24 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr7_-_4973960 | 0.24 |
ENSMUST00000144863.8
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr8_+_4399588 | 0.24 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr2_+_30331839 | 0.24 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr4_+_24898074 | 0.24 |
ENSMUST00000029925.10
ENSMUST00000151249.2 |
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr14_-_54936257 | 0.24 |
ENSMUST00000227947.2
ENSMUST00000224691.3 |
Gm29776
|
predicted gene, 29776 |
| chr1_+_21419819 | 0.24 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr11_-_42073737 | 0.24 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr19_+_6887059 | 0.23 |
ENSMUST00000088257.14
ENSMUST00000116551.10 |
Trmt112
|
tRNA methyltransferase 11-2 |
| chr2_+_30724984 | 0.23 |
ENSMUST00000113592.9
|
Prrx2
|
paired related homeobox 2 |
| chr7_+_103583554 | 0.23 |
ENSMUST00000214711.2
|
Olfr632
|
olfactory receptor 632 |
| chr10_-_87329513 | 0.23 |
ENSMUST00000020243.10
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr3_+_130904000 | 0.23 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr3_+_145926709 | 0.23 |
ENSMUST00000039164.4
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr17_-_28298721 | 0.23 |
ENSMUST00000233898.2
ENSMUST00000232798.2 |
Tcp11
|
t-complex protein 11 |
| chr7_-_106563137 | 0.23 |
ENSMUST00000213552.2
ENSMUST00000040983.6 ENSMUST00000213651.2 |
Olfr6
|
olfactory receptor 6 |
| chr2_-_3475990 | 0.23 |
ENSMUST00000060618.13
|
Suv39h2
|
suppressor of variegation 3-9 2 |
| chr11_+_70506674 | 0.23 |
ENSMUST00000180052.8
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr19_-_45800730 | 0.22 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr1_-_10079325 | 0.22 |
ENSMUST00000176398.8
ENSMUST00000027049.10 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr5_-_115332343 | 0.22 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr7_+_142086749 | 0.22 |
ENSMUST00000038675.7
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr5_-_124492734 | 0.22 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr7_-_134100659 | 0.22 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr13_-_53627110 | 0.22 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr16_-_44153498 | 0.22 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_+_70506716 | 0.22 |
ENSMUST00000144960.2
|
4930544D05Rik
|
RIKEN cDNA 4930544D05 gene |
| chr18_-_20135139 | 0.22 |
ENSMUST00000115848.5
|
Dsc3
|
desmocollin 3 |
| chr5_+_115568638 | 0.21 |
ENSMUST00000131079.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chr7_+_6733684 | 0.21 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr8_-_105036664 | 0.21 |
ENSMUST00000160596.8
ENSMUST00000164175.2 |
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr17_-_56416799 | 0.21 |
ENSMUST00000002908.8
ENSMUST00000238734.2 ENSMUST00000190703.7 |
Plin4
|
perilipin 4 |
| chr14_+_65835995 | 0.21 |
ENSMUST00000150897.8
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr17_-_57289121 | 0.21 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr19_+_6887486 | 0.21 |
ENSMUST00000174786.2
|
Trmt112
|
tRNA methyltransferase 11-2 |
| chr15_-_82505132 | 0.21 |
ENSMUST00000109515.3
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr1_+_171723231 | 0.21 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr2_-_151510453 | 0.20 |
ENSMUST00000180195.8
ENSMUST00000096439.4 |
Rad21l
|
RAD21-like (S. pombe) |
| chr8_-_105036739 | 0.20 |
ENSMUST00000159039.2
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chrX_-_139714182 | 0.20 |
ENSMUST00000044179.8
|
Tex13b
|
testis expressed 13B |
| chr1_+_78488440 | 0.20 |
ENSMUST00000152111.2
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr8_+_78939920 | 0.20 |
ENSMUST00000117845.8
ENSMUST00000126172.8 ENSMUST00000049395.15 |
Ttc29
|
tetratricopeptide repeat domain 29 |
| chr6_-_113694633 | 0.20 |
ENSMUST00000204533.3
|
Ghrl
|
ghrelin |
| chr16_+_17712061 | 0.20 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr7_+_140427729 | 0.19 |
ENSMUST00000106049.2
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr1_+_75522902 | 0.19 |
ENSMUST00000124341.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr17_+_35454833 | 0.19 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr7_+_5023375 | 0.19 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr9_-_20553576 | 0.19 |
ENSMUST00000155301.8
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr11_+_77409392 | 0.19 |
ENSMUST00000147386.2
|
Abhd15
|
abhydrolase domain containing 15 |
| chr19_-_46314945 | 0.19 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_+_140427711 | 0.19 |
ENSMUST00000026555.12
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr12_+_113038098 | 0.19 |
ENSMUST00000012355.14
ENSMUST00000146107.8 |
Tex22
|
testis expressed gene 22 |
| chr2_-_154411640 | 0.19 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr14_-_51134930 | 0.18 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr6_+_115921897 | 0.18 |
ENSMUST00000037831.14
ENSMUST00000161969.4 ENSMUST00000161617.7 |
H1f8
|
H1.8 linker histone |
| chr6_+_125169117 | 0.18 |
ENSMUST00000032485.7
|
Mrpl51
|
mitochondrial ribosomal protein L51 |
| chr10_-_21943978 | 0.18 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chr6_+_128991064 | 0.18 |
ENSMUST00000204981.2
|
Clec2f
|
C-type lectin domain family 2, member f |
| chr2_+_91757594 | 0.18 |
ENSMUST00000045537.4
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chrX_-_72442342 | 0.18 |
ENSMUST00000180787.3
|
Gm18336
|
predicted gene, 18336 |
| chr12_+_119278005 | 0.18 |
ENSMUST00000222784.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr12_+_113038376 | 0.18 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chrX_+_36675081 | 0.18 |
ENSMUST00000115179.4
|
Rhox2d
|
reproductive homeobox 2D |
| chr1_+_75213082 | 0.17 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chrX_+_36712105 | 0.17 |
ENSMUST00000072167.10
ENSMUST00000184746.2 |
Rhox2e
|
reproductive homeobox 2E |
| chr4_+_129941696 | 0.17 |
ENSMUST00000142293.8
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chrX_+_71025128 | 0.17 |
ENSMUST00000114569.2
|
Fate1
|
fetal and adult testis expressed 1 |
| chr10_+_100171538 | 0.17 |
ENSMUST00000186197.3
|
Gm4301
|
predicted gene 4301 |
| chr14_-_70879694 | 0.17 |
ENSMUST00000227123.2
ENSMUST00000022697.7 |
Fgf17
|
fibroblast growth factor 17 |
| chr18_+_48178397 | 0.17 |
ENSMUST00000076155.6
|
Eno1b
|
enolase 1B, retrotransposed |
| chr17_+_7246289 | 0.16 |
ENSMUST00000179728.2
|
Rnaset2b
|
ribonuclease T2B |
| chr6_-_52168675 | 0.16 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr18_+_38551960 | 0.16 |
ENSMUST00000236085.2
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_+_43097460 | 0.16 |
ENSMUST00000095725.11
|
Pdss2
|
prenyl (solanesyl) diphosphate synthase, subunit 2 |
| chr10_-_18662526 | 0.15 |
ENSMUST00000216654.2
|
Gm4922
|
predicted gene 4922 |
| chr11_-_69768875 | 0.15 |
ENSMUST00000178597.3
|
Tmem95
|
transmembrane protein 95 |
| chr17_+_33651864 | 0.15 |
ENSMUST00000174088.3
|
Actl9
|
actin-like 9 |
| chr1_+_75213044 | 0.15 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr17_+_34544632 | 0.15 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr3_-_46402355 | 0.15 |
ENSMUST00000195537.2
ENSMUST00000166505.7 ENSMUST00000195436.2 |
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr14_-_62693735 | 0.15 |
ENSMUST00000165651.8
ENSMUST00000022501.10 |
Gucy1b2
|
guanylate cyclase 1, soluble, beta 2 |
| chr2_-_173118315 | 0.15 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_134864731 | 0.14 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr14_-_78326435 | 0.14 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr16_-_18885809 | 0.14 |
ENSMUST00000200211.2
|
Iglj3
|
immunoglobulin lambda joining 3 |
| chr19_+_55730488 | 0.14 |
ENSMUST00000111659.9
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_-_51134906 | 0.14 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr15_-_76259126 | 0.14 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr13_-_48779072 | 0.14 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr7_+_6048160 | 0.14 |
ENSMUST00000037728.13
ENSMUST00000121583.2 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr10_+_129153986 | 0.14 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr7_-_126398165 | 0.13 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr11_+_120839288 | 0.13 |
ENSMUST00000070653.13
|
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr2_-_152672535 | 0.13 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr7_-_78432774 | 0.13 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr17_+_7246365 | 0.13 |
ENSMUST00000232245.2
|
Rnaset2b
|
ribonuclease T2B |
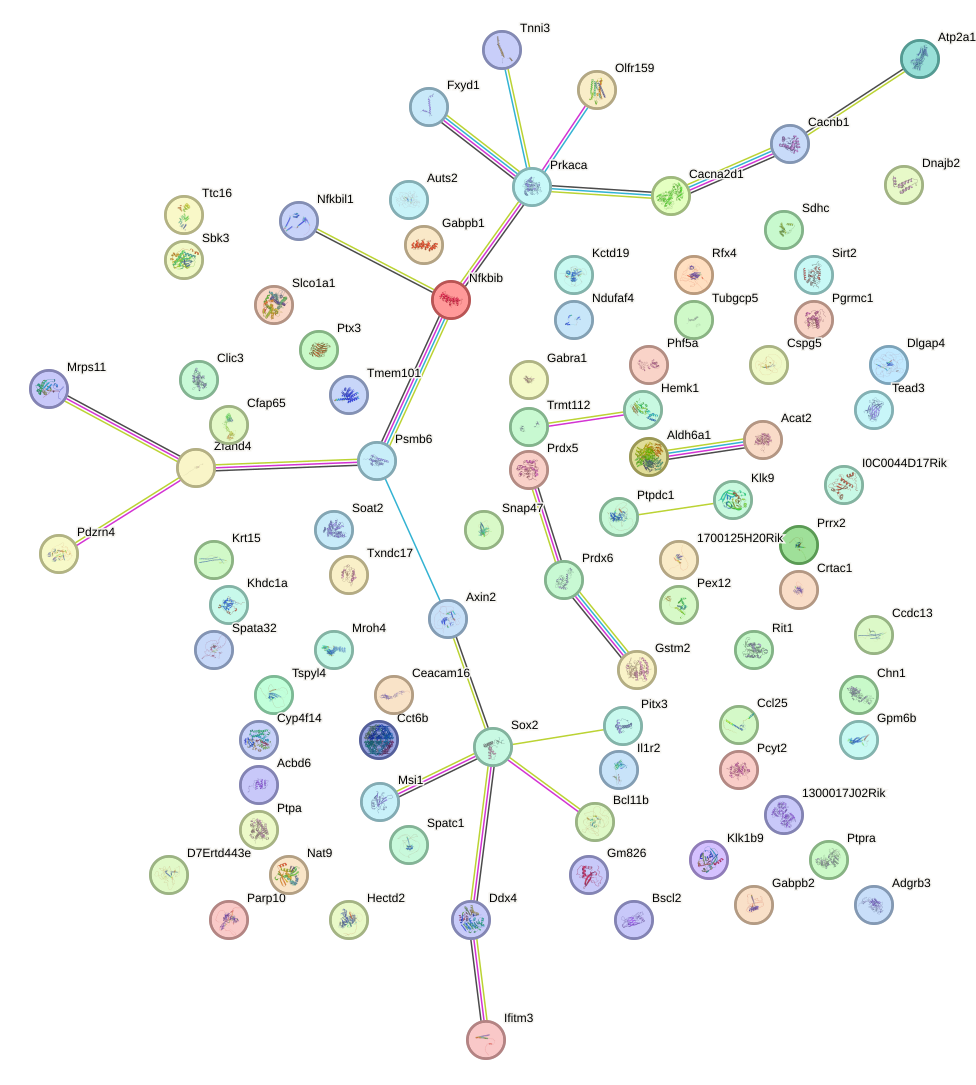
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 1.5 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 | 2.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 0.6 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 1.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.4 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.7 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.3 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.1 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 3.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.3 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) olfactory pit development(GO:0060166) |
| 0.1 | 0.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.6 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.8 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.5 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.1 | 0.2 | GO:0061152 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.1 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.2 | GO:1904349 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.6 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.7 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0060032 | floor plate formation(GO:0021508) notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 2.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.7 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.4 | 1.5 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 1.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 3.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.5 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 1.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |