Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
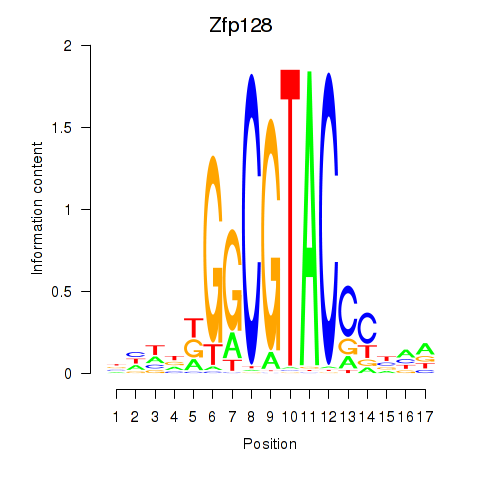
Results for Zfp128
Z-value: 0.66
Transcription factors associated with Zfp128
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp128
|
ENSMUSG00000060397.7 | zinc finger protein 128 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp128 | mm39_v1_chr7_+_12615091_12615132 | 0.49 | 2.4e-03 | Click! |
Activity profile of Zfp128 motif
Sorted Z-values of Zfp128 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_45175570 | 8.71 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr11_-_11920540 | 5.25 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_-_138169509 | 4.57 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138169253 | 4.43 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_128339882 | 4.00 |
ENSMUST00000073316.13
|
Foxm1
|
forkhead box M1 |
| chrX_-_133483849 | 3.82 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr13_-_59917569 | 2.92 |
ENSMUST00000057115.7
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr11_+_104441489 | 2.91 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_-_128339775 | 2.53 |
ENSMUST00000112152.8
ENSMUST00000057421.15 ENSMUST00000112151.2 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr5_-_138169476 | 2.46 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr15_-_96540760 | 2.44 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr7_+_45084257 | 2.26 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr8_+_84441806 | 2.25 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr8_+_84442133 | 2.11 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr4_+_126450728 | 2.04 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr15_-_76554065 | 1.91 |
ENSMUST00000037824.6
|
Foxh1
|
forkhead box H1 |
| chr7_-_44578834 | 1.85 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr7_+_45084300 | 1.84 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr8_+_84441854 | 1.83 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr7_-_45084012 | 1.72 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr8_-_127672590 | 1.16 |
ENSMUST00000179857.3
ENSMUST00000212771.2 |
Tomm20
|
translocase of outer mitochondrial membrane 20 |
| chr9_+_21323120 | 1.12 |
ENSMUST00000002902.8
|
Qtrt1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
| chr3_+_37474422 | 1.12 |
ENSMUST00000029277.13
ENSMUST00000198968.2 |
Spata5
|
spermatogenesis associated 5 |
| chr19_-_47525437 | 1.11 |
ENSMUST00000182808.8
ENSMUST00000182714.2 ENSMUST00000049369.16 |
Stn1
|
STN1, CST complex subunit |
| chr9_-_35087348 | 1.11 |
ENSMUST00000119847.2
ENSMUST00000034539.12 |
Dcps
|
decapping enzyme, scavenger |
| chr2_-_126985226 | 1.06 |
ENSMUST00000110386.2
ENSMUST00000132773.2 |
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
| chr14_-_106134253 | 0.95 |
ENSMUST00000022709.6
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr4_+_138960687 | 0.95 |
ENSMUST00000042675.9
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr12_-_75596441 | 0.83 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr6_-_128339458 | 0.75 |
ENSMUST00000155573.3
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr18_-_37777238 | 0.75 |
ENSMUST00000066272.6
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr17_+_34062059 | 0.73 |
ENSMUST00000002379.15
|
Cd320
|
CD320 antigen |
| chr2_-_30014466 | 0.72 |
ENSMUST00000044751.14
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr2_-_30014538 | 0.71 |
ENSMUST00000113677.3
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr9_+_3335469 | 0.66 |
ENSMUST00000053407.13
ENSMUST00000211933.2 ENSMUST00000212666.2 |
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr1_+_118409769 | 0.59 |
ENSMUST00000191823.6
|
Clasp1
|
CLIP associating protein 1 |
| chr15_+_76763431 | 0.47 |
ENSMUST00000023179.7
|
Zfp7
|
zinc finger protein 7 |
| chr11_-_116164928 | 0.40 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr3_+_97565528 | 0.38 |
ENSMUST00000045743.13
|
Prkab2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr11_-_116165024 | 0.37 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr5_-_121329385 | 0.34 |
ENSMUST00000054547.9
ENSMUST00000100770.9 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr3_-_79475078 | 0.27 |
ENSMUST00000133154.7
|
Fnip2
|
folliculin interacting protein 2 |
| chr9_+_3335135 | 0.25 |
ENSMUST00000212294.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr17_-_82045800 | 0.20 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_+_45175754 | 0.18 |
ENSMUST00000211227.2
ENSMUST00000051810.15 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr2_+_70886073 | 0.16 |
ENSMUST00000154704.8
ENSMUST00000135357.8 ENSMUST00000064141.12 ENSMUST00000112159.9 ENSMUST00000102701.5 |
Dcaf17
|
DDB1 and CUL4 associated factor 17 |
| chr7_-_21161866 | 0.03 |
ENSMUST00000177741.4
|
Gm6882
|
predicted gene 6882 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp128
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 1.3 | 3.8 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.0 | 11.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 1.9 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 | 5.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.4 | 1.9 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.4 | 1.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.4 | 2.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.3 | 1.7 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 1.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 4.0 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.2 | 1.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 1.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.3 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.9 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 3.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 3.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 2.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 1.4 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 8.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 1.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.4 | 1.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 1.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.9 | GO:0071203 | F-actin capping protein complex(GO:0008290) WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 4.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.4 | 1.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.3 | 2.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 13.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 1.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 8.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.8 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 5.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 1.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 8.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 8.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 5.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 4.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |