Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
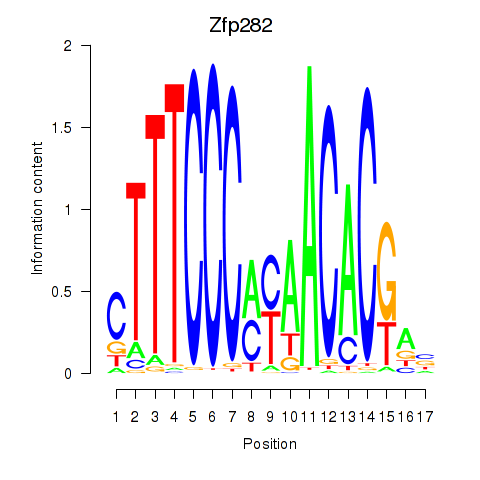
Results for Zfp282
Z-value: 0.72
Transcription factors associated with Zfp282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp282
|
ENSMUSG00000025821.10 | zinc finger protein 282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp282 | mm39_v1_chr6_+_47854138_47854138 | 0.76 | 1.0e-07 | Click! |
Activity profile of Zfp282 motif
Sorted Z-values of Zfp282 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_90576285 | 13.99 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr14_+_80237691 | 8.70 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr7_-_103492361 | 5.78 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr7_-_45175570 | 4.99 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_+_142558783 | 4.97 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr14_-_70864448 | 4.74 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_142558837 | 4.23 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chrX_+_134894573 | 3.21 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr8_+_95712151 | 3.20 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_114610879 | 3.00 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_+_134383247 | 2.84 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr6_-_83504471 | 2.51 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_24705320 | 2.40 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr6_-_83504756 | 2.35 |
ENSMUST00000152029.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr5_-_65548723 | 2.28 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chr8_-_23727639 | 2.13 |
ENSMUST00000033950.7
|
Gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr11_-_68864666 | 2.10 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr16_+_10884156 | 2.00 |
ENSMUST00000089011.6
|
Snn
|
stannin |
| chr13_-_95661726 | 1.97 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr9_+_120400510 | 1.85 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr10_+_93289264 | 1.84 |
ENSMUST00000016033.9
|
Lta4h
|
leukotriene A4 hydrolase |
| chr9_-_95727267 | 1.78 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr19_-_6835538 | 1.76 |
ENSMUST00000113440.2
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr17_-_50401305 | 1.76 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr1_+_39407183 | 1.44 |
ENSMUST00000195123.6
|
Rpl31
|
ribosomal protein L31 |
| chr1_-_125362321 | 1.42 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr10_-_19727165 | 1.41 |
ENSMUST00000059805.6
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr18_-_35855383 | 1.28 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr4_-_126096376 | 1.22 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr1_+_74448535 | 1.18 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr17_-_33904386 | 1.15 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr17_-_33904345 | 1.13 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr12_-_87490580 | 1.02 |
ENSMUST00000162961.8
ENSMUST00000185301.2 |
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr13_-_74882374 | 0.92 |
ENSMUST00000220738.2
|
Cast
|
calpastatin |
| chr7_-_105289515 | 0.91 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr4_-_126096551 | 0.89 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr4_+_14864076 | 0.89 |
ENSMUST00000029875.4
|
Pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr2_-_24985137 | 0.88 |
ENSMUST00000114373.8
|
Noxa1
|
NADPH oxidase activator 1 |
| chr2_-_24985161 | 0.86 |
ENSMUST00000044018.8
|
Noxa1
|
NADPH oxidase activator 1 |
| chr16_-_85599994 | 0.84 |
ENSMUST00000023610.15
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
| chr7_+_16472335 | 0.73 |
ENSMUST00000086112.8
ENSMUST00000205607.2 |
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr13_-_74882328 | 0.70 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr3_-_88858465 | 0.66 |
ENSMUST00000173135.8
|
Dap3
|
death associated protein 3 |
| chr3_-_88858402 | 0.63 |
ENSMUST00000173021.8
|
Dap3
|
death associated protein 3 |
| chrX_+_105964224 | 0.62 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr13_-_22289994 | 0.61 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr17_-_46991709 | 0.58 |
ENSMUST00000233524.2
ENSMUST00000233733.2 ENSMUST00000071841.7 ENSMUST00000165007.9 |
Klhdc3
|
kelch domain containing 3 |
| chr12_+_72583114 | 0.56 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr10_-_41894360 | 0.45 |
ENSMUST00000162405.8
ENSMUST00000095729.11 ENSMUST00000161081.2 ENSMUST00000160262.9 |
Armc2
|
armadillo repeat containing 2 |
| chr2_-_125466985 | 0.45 |
ENSMUST00000089776.3
|
Cep152
|
centrosomal protein 152 |
| chr7_-_143294051 | 0.41 |
ENSMUST00000119499.8
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr12_+_111504450 | 0.35 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr10_-_51507527 | 0.26 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr18_-_78222349 | 0.25 |
ENSMUST00000237290.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr5_-_87638728 | 0.23 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr16_+_7011580 | 0.22 |
ENSMUST00000231194.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_+_35011953 | 0.22 |
ENSMUST00000069837.4
|
Slit3
|
slit guidance ligand 3 |
| chr9_-_85631361 | 0.21 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_+_82933383 | 0.18 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr17_-_47169380 | 0.17 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr2_-_91274967 | 0.17 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr12_+_9624437 | 0.17 |
ENSMUST00000057021.9
|
Osr1
|
odd-skipped related transcription factor 1 |
| chr7_-_102507962 | 0.16 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr7_-_35255729 | 0.15 |
ENSMUST00000040962.6
|
Nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr19_-_5135510 | 0.15 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr7_-_29772226 | 0.13 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr19_-_5538370 | 0.13 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr7_-_12552764 | 0.13 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr12_+_111504640 | 0.13 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr12_+_87490666 | 0.12 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr16_-_34334454 | 0.11 |
ENSMUST00000089655.12
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_+_24276545 | 0.09 |
ENSMUST00000127242.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr11_-_102120953 | 0.09 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr2_+_25179903 | 0.08 |
ENSMUST00000028337.7
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr7_+_26456567 | 0.08 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr11_-_102120917 | 0.08 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr17_+_47221377 | 0.07 |
ENSMUST00000024773.6
|
Prph2
|
peripherin 2 |
| chr19_+_56814700 | 0.05 |
ENSMUST00000078723.11
ENSMUST00000121249.8 |
Tdrd1
|
tudor domain containing 1 |
| chr2_+_28386895 | 0.03 |
ENSMUST00000163121.8
|
Gbgt1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 |
| chr7_-_46569662 | 0.03 |
ENSMUST00000143413.3
ENSMUST00000014546.15 |
Tsg101
|
tumor susceptibility gene 101 |
| chr2_+_37666243 | 0.03 |
ENSMUST00000050372.10
|
Crb2
|
crumbs family member 2 |
| chr6_-_39787813 | 0.03 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr7_-_108484213 | 0.02 |
ENSMUST00000217803.2
|
Olfr518
|
olfactory receptor 518 |
| chr16_+_87251852 | 0.01 |
ENSMUST00000119504.8
ENSMUST00000131356.8 |
Usp16
|
ubiquitin specific peptidase 16 |
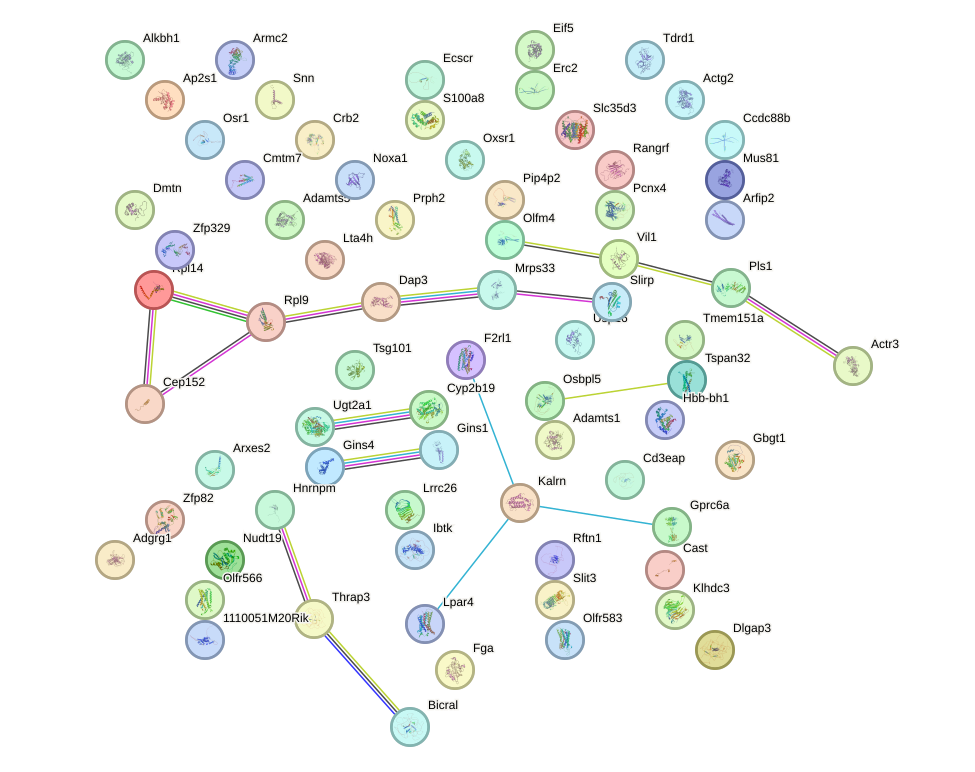
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp282
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.6 | 4.7 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.8 | 9.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.8 | 4.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 3.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.7 | 3.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.7 | 2.0 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.6 | 5.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 1.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 2.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 2.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 1.4 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.3 | 1.0 | GO:0042245 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.2 | 2.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 1.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 2.4 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 8.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 3.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.7 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 1.8 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:0072194 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 1.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.8 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 3.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 3.9 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 1.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.7 | 5.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.7 | 2.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.7 | 4.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 8.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 5.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 3.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 2.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 14.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 5.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.4 | 5.8 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.5 | 2.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 1.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 1.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 2.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.0 | GO:0070579 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 1.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 5.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 4.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 3.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 6.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 8.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.0 | GO:0005125 | cytokine activity(GO:0005125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 5.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |