Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zfp410
Z-value: 0.81
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.12 | zinc finger protein 410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp410 | mm39_v1_chr12_+_84363876_84363920 | 0.78 | 2.8e-08 | Click! |
Activity profile of Zfp410 motif
Sorted Z-values of Zfp410 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_125072944 | 14.16 |
ENSMUST00000112392.8
ENSMUST00000056889.15 |
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr6_+_125073108 | 9.69 |
ENSMUST00000112390.8
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr15_-_89310060 | 5.62 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr14_-_52434863 | 2.24 |
ENSMUST00000046709.9
|
Supt16
|
SPT16, facilitates chromatin remodeling subunit |
| chr15_-_79718423 | 1.97 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr6_+_68013772 | 1.89 |
ENSMUST00000197515.2
ENSMUST00000103315.3 |
Igkv17-121
|
immunoglobulin kappa variable 17-121 |
| chr14_-_56299764 | 1.65 |
ENSMUST00000043249.10
|
Mcpt4
|
mast cell protease 4 |
| chr6_-_142418801 | 1.43 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr6_+_67838100 | 1.41 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chrX_+_133618693 | 1.18 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr16_-_21606546 | 1.03 |
ENSMUST00000023559.7
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr5_+_123280250 | 0.99 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr11_+_22462088 | 0.74 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr5_-_100648487 | 0.54 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr5_-_104125226 | 0.35 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_-_131455039 | 0.26 |
ENSMUST00000097588.9
ENSMUST00000186543.7 |
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_-_104125270 | 0.23 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_55982969 | 0.20 |
ENSMUST00000063138.8
|
Msl3l2
|
MSL3 like 2 |
| chr5_-_104125192 | 0.09 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
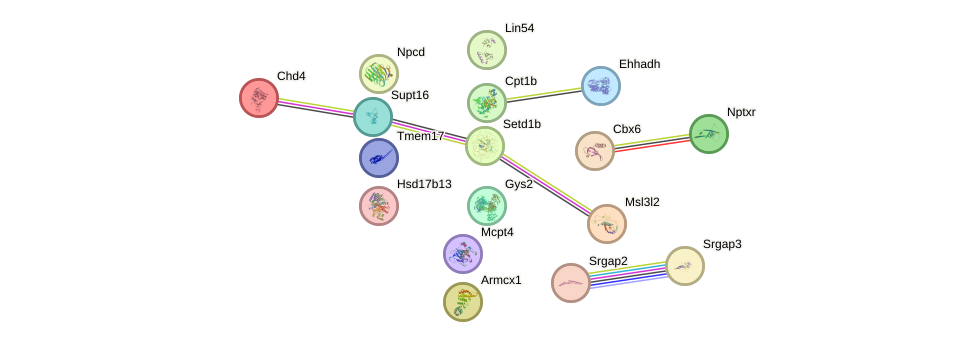
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp410
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 23.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 1.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 2.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 5.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 1.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 3.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 23.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.6 | 2.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 5.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 23.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.5 | 1.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 2.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 23.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 5.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |