Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
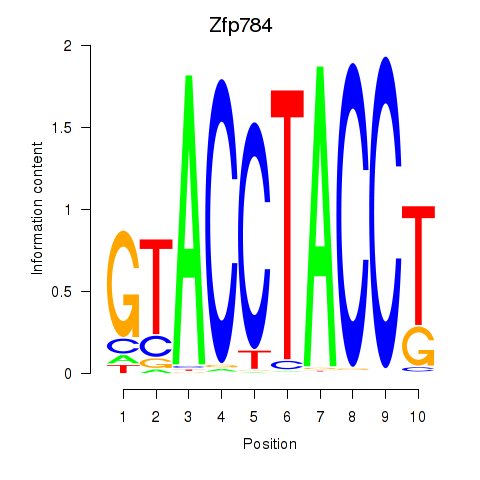
Results for Zfp784
Z-value: 0.64
Transcription factors associated with Zfp784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp784
|
ENSMUSG00000043290.7 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp784 | mm39_v1_chr7_-_5041427_5041450 | 0.53 | 9.1e-04 | Click! |
Activity profile of Zfp784 motif
Sorted Z-values of Zfp784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_130119077 | 4.12 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr7_-_103463120 | 3.97 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr14_-_47655621 | 3.41 |
ENSMUST00000180299.8
|
Dlgap5
|
DLG associated protein 5 |
| chrX_-_138772383 | 3.33 |
ENSMUST00000033811.14
ENSMUST00000087401.12 |
Morc4
|
microrchidia 4 |
| chr1_-_133681419 | 3.32 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr13_+_73615316 | 3.31 |
ENSMUST00000022099.15
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr14_-_70864448 | 3.25 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr15_-_78456898 | 3.02 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr7_-_45173193 | 2.95 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_+_142025817 | 2.84 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr2_+_118644717 | 2.81 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr7_+_142025575 | 2.73 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr14_-_70864666 | 2.70 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr6_+_86055018 | 2.69 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr11_-_101979297 | 2.69 |
ENSMUST00000017458.11
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr2_+_118644675 | 2.43 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr3_+_146110387 | 2.42 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr12_+_24758240 | 2.40 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr8_-_71219299 | 2.32 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr17_+_12338161 | 2.20 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr13_-_100912308 | 2.15 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr2_+_118644475 | 2.11 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr6_+_86055048 | 2.07 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chrX_-_149372840 | 1.95 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr7_-_110462446 | 1.89 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr9_-_20864096 | 1.86 |
ENSMUST00000004202.17
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr4_-_152561896 | 1.71 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr4_-_87951565 | 1.67 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr4_-_63090355 | 1.67 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chr6_+_86605146 | 1.63 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr12_-_79054050 | 1.60 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr2_+_124910037 | 1.58 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr1_+_151447124 | 1.58 |
ENSMUST00000148810.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr10_-_127031578 | 1.57 |
ENSMUST00000038217.14
ENSMUST00000130855.8 ENSMUST00000116229.2 ENSMUST00000144322.8 |
Dtx3
|
deltex 3, E3 ubiquitin ligase |
| chr13_+_52750883 | 1.55 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr11_+_69855584 | 1.55 |
ENSMUST00000108597.8
ENSMUST00000060651.6 ENSMUST00000108596.8 |
Cldn7
|
claudin 7 |
| chr8_-_121316043 | 1.51 |
ENSMUST00000034278.6
|
Gins2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr7_+_120442773 | 1.50 |
ENSMUST00000143279.3
ENSMUST00000106489.9 |
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr11_+_75542902 | 1.49 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr5_+_123887759 | 1.49 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr7_-_27242170 | 1.43 |
ENSMUST00000150964.2
|
Pld3
|
phospholipase D family, member 3 |
| chr19_+_4204605 | 1.39 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr11_-_34674677 | 1.37 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr3_-_54642450 | 1.35 |
ENSMUST00000153224.2
|
Exosc8
|
exosome component 8 |
| chr2_-_38895586 | 1.32 |
ENSMUST00000080861.6
|
Rpl35
|
ribosomal protein L35 |
| chr7_-_24997393 | 1.31 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr13_+_20978283 | 1.31 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr16_+_36755338 | 1.28 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr7_+_120442048 | 1.26 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr7_-_24705320 | 1.20 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr12_+_113112311 | 1.19 |
ENSMUST00000199089.5
|
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr2_+_24852409 | 1.18 |
ENSMUST00000028351.9
|
Dph7
|
diphthamine biosynethesis 7 |
| chr3_-_37778470 | 1.18 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chrX_-_132799041 | 1.17 |
ENSMUST00000176718.8
ENSMUST00000176641.2 |
Tspan6
|
tetraspanin 6 |
| chr8_+_105067159 | 1.16 |
ENSMUST00000212948.2
ENSMUST00000034343.5 |
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr5_-_137609634 | 1.15 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr11_+_75542328 | 1.14 |
ENSMUST00000069057.13
|
Myo1c
|
myosin IC |
| chr5_+_114268425 | 1.13 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr17_-_23959334 | 1.09 |
ENSMUST00000024702.5
|
Paqr4
|
progestin and adipoQ receptor family member IV |
| chr9_-_119897328 | 1.09 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr5_-_137609691 | 1.08 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr4_+_129407374 | 1.08 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr9_-_119897358 | 1.06 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chrX_+_154045439 | 1.06 |
ENSMUST00000026324.10
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr16_+_35590745 | 1.03 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr6_-_124710030 | 1.02 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_-_24997291 | 1.02 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr16_+_4756963 | 1.02 |
ENSMUST00000229961.2
ENSMUST00000050881.10 |
Nudt16l1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
| chr11_+_50267808 | 0.97 |
ENSMUST00000109142.8
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr8_+_124750133 | 0.92 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr14_-_61275340 | 0.90 |
ENSMUST00000225730.2
ENSMUST00000111236.4 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr17_-_13211374 | 0.88 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr5_-_99876671 | 0.87 |
ENSMUST00000209346.2
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr6_-_72357398 | 0.87 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr3_+_146110709 | 0.86 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr7_-_48530777 | 0.86 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr4_-_154059495 | 0.85 |
ENSMUST00000030893.3
|
Dffb
|
DNA fragmentation factor, beta subunit |
| chrX_-_135644424 | 0.84 |
ENSMUST00000166478.8
ENSMUST00000113097.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr4_-_59783780 | 0.84 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr17_-_48235385 | 0.84 |
ENSMUST00000113262.2
|
Foxp4
|
forkhead box P4 |
| chr17_-_48235560 | 0.83 |
ENSMUST00000113265.8
|
Foxp4
|
forkhead box P4 |
| chr2_-_165125519 | 0.82 |
ENSMUST00000155289.8
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr8_+_105066924 | 0.82 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr2_+_84867783 | 0.81 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr4_-_130068484 | 0.80 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr16_-_18107046 | 0.80 |
ENSMUST00000232424.2
ENSMUST00000009321.11 |
Dgcr8
|
DGCR8, microprocessor complex subunit |
| chr17_-_48235325 | 0.79 |
ENSMUST00000113263.8
ENSMUST00000097311.9 |
Foxp4
|
forkhead box P4 |
| chr16_+_20536856 | 0.79 |
ENSMUST00000231392.2
ENSMUST00000161038.2 |
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr12_-_40298072 | 0.78 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr2_+_118219096 | 0.78 |
ENSMUST00000110877.8
ENSMUST00000005233.12 |
Eif2ak4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr13_+_107083613 | 0.76 |
ENSMUST00000022203.10
|
Dimt1
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr8_-_13155431 | 0.76 |
ENSMUST00000164416.8
|
Pcid2
|
PCI domain containing 2 |
| chr5_+_64969679 | 0.75 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr11_+_77923172 | 0.75 |
ENSMUST00000122342.2
ENSMUST00000092881.4 |
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr6_-_124710084 | 0.75 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_-_136031799 | 0.73 |
ENSMUST00000196159.5
ENSMUST00000041577.13 |
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr4_+_135639117 | 0.72 |
ENSMUST00000068830.4
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chr15_+_34495441 | 0.71 |
ENSMUST00000052290.14
ENSMUST00000079028.6 |
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
| chr3_+_84859453 | 0.71 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr16_-_74208180 | 0.70 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr6_-_87798613 | 0.70 |
ENSMUST00000204169.2
|
Gm45140
|
predicted gene 45140 |
| chr16_-_3821614 | 0.70 |
ENSMUST00000171658.2
ENSMUST00000171762.2 |
Slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr4_-_92079986 | 0.69 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chr17_-_13211075 | 0.69 |
ENSMUST00000159986.8
ENSMUST00000007007.14 |
Wtap
|
Wilms tumour 1-associating protein |
| chrX_+_20291927 | 0.69 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr7_+_25327028 | 0.67 |
ENSMUST00000076034.8
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_+_151447232 | 0.67 |
ENSMUST00000111875.2
|
Niban1
|
niban apoptosis regulator 1 |
| chr11_+_68393845 | 0.66 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr12_+_55171254 | 0.66 |
ENSMUST00000177768.3
|
Fam177a
|
family with sequence similarity 177, member A |
| chr1_+_40363701 | 0.66 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr11_+_77923100 | 0.63 |
ENSMUST00000021187.12
|
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr4_+_154059619 | 0.63 |
ENSMUST00000047497.15
|
Cep104
|
centrosomal protein 104 |
| chr4_+_11558905 | 0.61 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr7_-_127529238 | 0.59 |
ENSMUST00000032988.10
ENSMUST00000206124.2 |
Prss8
|
protease, serine 8 (prostasin) |
| chr14_-_86986541 | 0.58 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr1_+_9868332 | 0.58 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr10_-_7556881 | 0.58 |
ENSMUST00000159977.2
ENSMUST00000162682.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr2_-_45003270 | 0.56 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_-_19019100 | 0.55 |
ENSMUST00000103749.3
|
Iglc2
|
immunoglobulin lambda constant 2 |
| chr15_+_102427149 | 0.54 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chrX_+_73372664 | 0.54 |
ENSMUST00000004326.4
|
Plxna3
|
plexin A3 |
| chr7_+_101583283 | 0.54 |
ENSMUST00000209639.2
ENSMUST00000210679.2 |
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr18_+_3507917 | 0.52 |
ENSMUST00000025075.3
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chr2_+_101455079 | 0.51 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr1_+_133965228 | 0.50 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr11_-_30218167 | 0.50 |
ENSMUST00000006629.14
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr17_+_6047112 | 0.49 |
ENSMUST00000115786.8
|
Synj2
|
synaptojanin 2 |
| chr7_-_108774367 | 0.49 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr3_+_88536749 | 0.49 |
ENSMUST00000176316.8
ENSMUST00000176879.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_-_5944098 | 0.47 |
ENSMUST00000055911.6
ENSMUST00000236767.2 |
Tigd3
|
tigger transposable element derived 3 |
| chr4_+_131649001 | 0.46 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr18_+_11766333 | 0.45 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr12_+_31315227 | 0.44 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr2_-_45002902 | 0.44 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_+_49835576 | 0.43 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr4_-_11007635 | 0.42 |
ENSMUST00000054776.4
|
Plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr13_+_21901791 | 0.42 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr12_-_46863726 | 0.42 |
ENSMUST00000219330.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr5_-_107873883 | 0.40 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr9_+_90045219 | 0.40 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr6_+_29361408 | 0.40 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr18_+_35932894 | 0.40 |
ENSMUST00000236484.2
ENSMUST00000236550.2 ENSMUST00000235919.2 |
Ube2d2a
Gm50371
|
ubiquitin-conjugating enzyme E2D 2A predicted gene, 50371 |
| chr3_-_136031723 | 0.40 |
ENSMUST00000198206.2
|
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr17_-_82045800 | 0.39 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr17_+_69569184 | 0.39 |
ENSMUST00000224951.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr1_+_171594690 | 0.38 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr15_-_103123711 | 0.38 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr4_-_4138432 | 0.38 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr11_+_6511133 | 0.38 |
ENSMUST00000160633.8
ENSMUST00000109721.3 |
Ccm2
|
cerebral cavernous malformation 2 |
| chr19_+_29344846 | 0.38 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen |
| chr9_+_90045109 | 0.38 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr11_+_97206542 | 0.38 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr13_+_49835667 | 0.38 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr2_+_101455022 | 0.38 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr13_+_55468313 | 0.36 |
ENSMUST00000021942.8
|
Prelid1
|
PRELI domain containing 1 |
| chr3_+_87837566 | 0.36 |
ENSMUST00000055984.7
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr18_+_34892599 | 0.36 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr2_+_18069375 | 0.36 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr5_-_112399003 | 0.36 |
ENSMUST00000112383.8
|
Cryba4
|
crystallin, beta A4 |
| chr19_+_55882942 | 0.35 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr13_-_20008397 | 0.34 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr18_-_68562385 | 0.32 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr11_-_62283431 | 0.31 |
ENSMUST00000151498.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr11_+_107438751 | 0.30 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr3_-_108117754 | 0.30 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr17_+_43671314 | 0.30 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr4_-_130068902 | 0.30 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr4_+_131648509 | 0.29 |
ENSMUST00000238733.2
|
Tmem200b
|
transmembrane protein 200B |
| chr5_+_135698881 | 0.29 |
ENSMUST00000153500.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr4_+_62327010 | 0.29 |
ENSMUST00000084524.4
|
Prpf4
|
pre-mRNA processing factor 4 |
| chr14_-_63482668 | 0.28 |
ENSMUST00000118022.8
|
Gata4
|
GATA binding protein 4 |
| chr15_+_82012114 | 0.28 |
ENSMUST00000089174.6
|
Ccdc134
|
coiled-coil domain containing 134 |
| chr8_-_112580910 | 0.28 |
ENSMUST00000034432.7
|
Cfdp1
|
craniofacial development protein 1 |
| chr2_+_174126103 | 0.27 |
ENSMUST00000109095.8
ENSMUST00000180362.8 ENSMUST00000109096.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr14_-_63482720 | 0.27 |
ENSMUST00000067417.10
|
Gata4
|
GATA binding protein 4 |
| chr9_+_19620729 | 0.27 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr13_+_48816466 | 0.27 |
ENSMUST00000021813.5
|
Barx1
|
BarH-like homeobox 1 |
| chr13_+_47347301 | 0.26 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr19_-_29344694 | 0.26 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr19_-_6964988 | 0.26 |
ENSMUST00000130048.8
ENSMUST00000025914.7 |
Vegfb
|
vascular endothelial growth factor B |
| chr8_+_105066980 | 0.25 |
ENSMUST00000211885.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr2_+_180222985 | 0.25 |
ENSMUST00000169630.8
|
Mrgbp
|
MRG/MORF4L binding protein |
| chr6_-_113354337 | 0.24 |
ENSMUST00000043333.9
|
Tada3
|
transcriptional adaptor 3 |
| chr7_+_18810097 | 0.22 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr8_-_46605196 | 0.22 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr19_-_40371016 | 0.22 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_-_146731810 | 0.22 |
ENSMUST00000085614.6
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr2_-_60552980 | 0.21 |
ENSMUST00000028348.9
ENSMUST00000112517.8 |
Itgb6
|
integrin beta 6 |
| chr10_-_30679071 | 0.21 |
ENSMUST00000068567.5
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr2_-_87504008 | 0.21 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr10_-_30679289 | 0.20 |
ENSMUST00000215725.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_+_106908709 | 0.20 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr2_+_152907875 | 0.19 |
ENSMUST00000238488.2
ENSMUST00000129377.8 ENSMUST00000109800.2 |
Ccm2l
|
cerebral cavernous malformation 2-like |
| chr17_+_44499451 | 0.19 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr5_+_122344854 | 0.18 |
ENSMUST00000145854.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr10_+_78412783 | 0.17 |
ENSMUST00000219588.2
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_+_30745447 | 0.17 |
ENSMUST00000066295.5
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr1_-_173569301 | 0.17 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr5_-_93192881 | 0.17 |
ENSMUST00000061328.6
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr9_-_60048503 | 0.17 |
ENSMUST00000034829.6
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
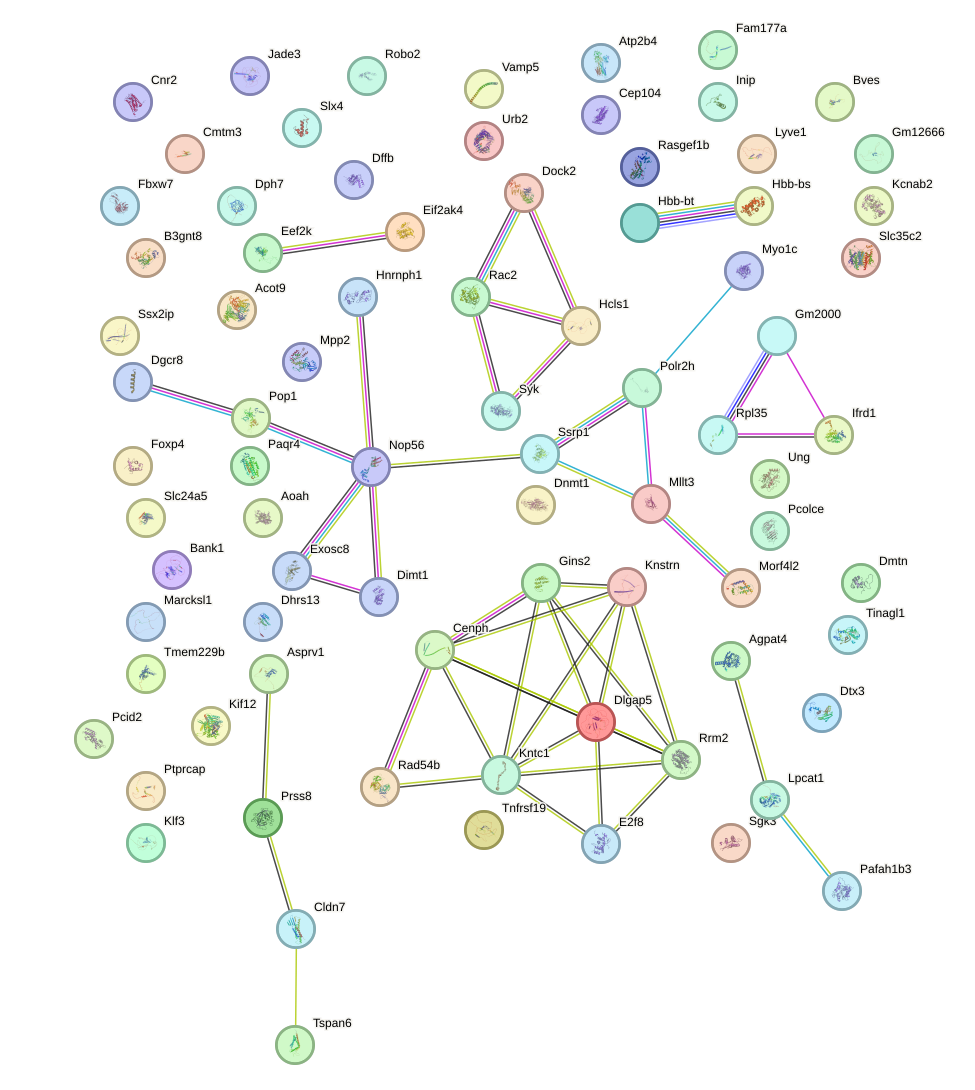
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.0 | 6.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 3.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.8 | 3.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.7 | 2.1 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.6 | 1.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.5 | 3.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.5 | 1.9 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 1.4 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.4 | 1.6 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.4 | 1.5 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.4 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.3 | 0.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 | 7.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 2.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 3.3 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 0.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 0.8 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 0.8 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.2 | 0.7 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.2 | 0.7 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 2.5 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 | 1.8 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 2.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 0.8 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.2 | 2.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.8 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 0.6 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.2 | 0.9 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.2 | 0.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.5 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 4.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 4.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 1.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 1.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 2.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.4 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 1.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 1.6 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0050689 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 1.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.8 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.1 | 0.3 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.5 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 | 0.3 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.5 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.3 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 0.7 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.7 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.3 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.5 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.4 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.6 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.4 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 1.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 2.5 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.6 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 4.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 1.7 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.5 | 2.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 1.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.5 | 1.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 4.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 2.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 4.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 0.8 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 1.6 | GO:0036396 | MIS complex(GO:0036396) |
| 0.2 | 3.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 7.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 3.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 3.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 3.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 2.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.0 | 4.0 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.9 | 2.8 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.5 | 1.9 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.4 | 2.4 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 2.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 1.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 0.8 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.2 | 2.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 0.7 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 4.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 3.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 1.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 1.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 0.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.8 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.7 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 1.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 2.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 9.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.0 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 1.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 2.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.1 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0051430 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 3.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |