Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
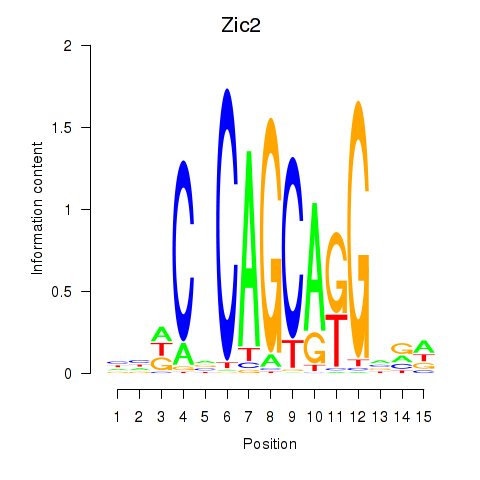
Results for Zic2
Z-value: 1.16
Transcription factors associated with Zic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic2
|
ENSMUSG00000061524.9 | zinc finger protein of the cerebellum 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic2 | mm39_v1_chr14_+_122712809_122712847 | 0.14 | 4.2e-01 | Click! |
Activity profile of Zic2 motif
Sorted Z-values of Zic2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.6 | 10.3 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 2.4 | 7.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 1.9 | 5.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.7 | 5.1 | GO:0010924 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 1.7 | 5.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 1.4 | 4.3 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.4 | 1.4 | GO:0072313 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 1.4 | 6.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 1.2 | 3.5 | GO:0045212 | acetylcholine metabolic process(GO:0008291) neurotransmitter receptor biosynthetic process(GO:0045212) acetate ester metabolic process(GO:1900619) |
| 1.1 | 4.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.8 | 3.4 | GO:0090290 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.8 | 2.5 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 0.8 | 6.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.7 | 2.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.7 | 2.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.7 | 0.7 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.6 | 3.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 | 7.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.6 | 2.4 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.6 | 1.8 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.6 | 7.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 4.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.5 | 3.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.5 | 1.5 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-10 biosynthetic process(GO:0045074) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.5 | 1.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.5 | 1.9 | GO:0061188 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.5 | 1.8 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.5 | 4.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 2.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.5 | 0.9 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.4 | 1.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 4.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.4 | 1.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.4 | 2.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 3.7 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.4 | 1.4 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.4 | 2.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.4 | 3.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 5.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.3 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.3 | 3.6 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.3 | 0.6 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.3 | 1.8 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.3 | 2.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 1.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 2.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 3.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.3 | 3.7 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 1.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 3.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.3 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 1.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 3.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.7 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 5.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 0.7 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.2 | 1.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 2.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 5.6 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 1.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 2.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.2 | 1.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 0.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 2.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 3.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 0.9 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 0.6 | GO:0042495 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.2 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 2.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.7 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 1.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 1.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 4.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.9 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 1.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 1.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 2.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 2.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.3 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.1 | 1.8 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 0.5 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 0.7 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:0061228 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.7 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.1 | 1.9 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.6 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 6.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.9 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 3.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.6 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.7 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 1.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 2.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.6 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 1.0 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 | 1.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 2.5 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.1 | 1.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.9 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.3 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 5.0 | GO:1902850 | microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 0.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 2.3 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 1.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.9 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 6.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 1.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.6 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 0.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.3 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.7 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 4.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 1.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 1.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 2.8 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 2.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 1.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 2.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 3.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.5 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 1.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 2.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 1.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 4.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.6 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 2.4 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.9 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 1.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.3 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0003219 | secondary heart field specification(GO:0003139) cardiac right ventricle formation(GO:0003219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.6 | 4.8 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.5 | 4.6 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.0 | 3.9 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.9 | 2.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 5.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 3.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 6.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 4.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 3.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 2.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 2.9 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.3 | 1.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 5.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.3 | 3.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 1.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 7.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 3.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 3.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 1.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.9 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 4.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.7 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 14.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 3.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 3.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.3 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.7 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 5.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 2.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 5.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 2.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 5.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 5.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 3.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.4 | 5.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.2 | 3.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.0 | 2.9 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.9 | 4.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.7 | 2.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.7 | 12.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.6 | 1.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.6 | 2.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 1.6 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.5 | 1.9 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 5.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 2.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 1.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.4 | 5.0 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.4 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.4 | 1.4 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.3 | 10.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 6.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 23.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 2.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 0.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 0.9 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.3 | 0.9 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.3 | 2.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.3 | 2.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 3.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 0.7 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 0.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 2.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 4.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 2.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 1.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 2.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 2.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.6 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 1.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 1.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.7 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 2.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 2.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 3.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 5.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.9 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 1.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.9 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 3.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.6 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 1.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 3.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 6.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 3.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 3.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 9.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 3.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 3.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 4.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 3.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.1 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.2 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 4.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 3.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 5.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 4.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 4.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 4.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 6.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 14.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 12.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 3.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 6.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 5.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 7.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 10.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 6.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 5.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 5.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 5.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 5.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 10.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 4.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 5.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 1.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 3.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 2.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 4.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 6.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 10.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 4.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 5.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 3.9 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |