Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
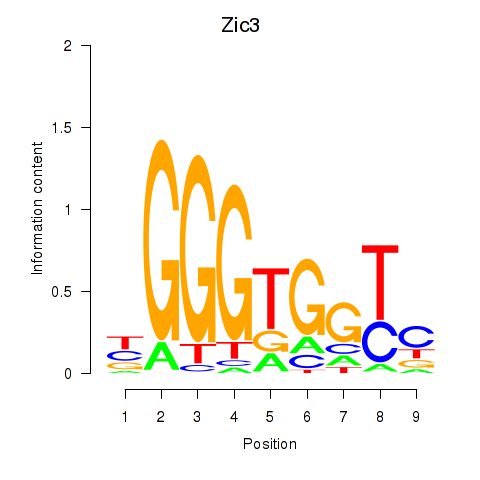
Results for Zic3
Z-value: 0.56
Transcription factors associated with Zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic3
|
ENSMUSG00000067860.12 | zinc finger protein of the cerebellum 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | mm39_v1_chrX_+_57076359_57076378 | -0.05 | 7.5e-01 | Click! |
Activity profile of Zic3 motif
Sorted Z-values of Zic3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_24069680 | 2.37 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr11_+_115790768 | 1.73 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr12_-_76756772 | 1.71 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr7_-_16796309 | 1.70 |
ENSMUST00000153833.2
ENSMUST00000108492.9 |
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr18_-_78185334 | 1.68 |
ENSMUST00000160639.2
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr11_-_85030761 | 1.58 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32 |
| chrX_+_55500170 | 1.32 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr1_+_82817794 | 1.31 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr1_+_82817388 | 1.20 |
ENSMUST00000190052.7
ENSMUST00000063380.11 ENSMUST00000187899.7 ENSMUST00000186302.7 ENSMUST00000190046.7 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr11_+_115790951 | 1.20 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr2_-_170269748 | 1.11 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr1_-_133728779 | 1.11 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr2_+_29759495 | 1.09 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr8_-_86091946 | 1.07 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_-_107960528 | 1.01 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr13_+_6598185 | 0.95 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr11_-_90578397 | 0.95 |
ENSMUST00000107869.9
ENSMUST00000154599.2 ENSMUST00000107868.8 ENSMUST00000020849.9 |
Tom1l1
|
target of myb1-like 1 (chicken) |
| chr14_-_73563212 | 0.92 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr11_+_69806866 | 0.89 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr7_+_110372860 | 0.89 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chrX_+_139857688 | 0.88 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr19_-_5776268 | 0.86 |
ENSMUST00000075606.6
ENSMUST00000236215.2 ENSMUST00000235730.2 ENSMUST00000237081.2 ENSMUST00000049295.15 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chrX_+_139857640 | 0.83 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr2_+_78699360 | 0.79 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr4_+_52439237 | 0.79 |
ENSMUST00000102915.10
ENSMUST00000117280.8 ENSMUST00000142227.3 |
Smc2
|
structural maintenance of chromosomes 2 |
| chr7_+_78563964 | 0.76 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr2_+_29780122 | 0.75 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr10_+_82821304 | 0.75 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr11_+_53410697 | 0.74 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr14_+_30853010 | 0.70 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr11_-_4544751 | 0.68 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr17_-_24428351 | 0.68 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr13_+_81931196 | 0.67 |
ENSMUST00000022009.10
ENSMUST00000223793.2 |
Cetn3
|
centrin 3 |
| chr5_+_34731152 | 0.67 |
ENSMUST00000001108.11
ENSMUST00000114340.9 |
Add1
|
adducin 1 (alpha) |
| chr5_+_34731008 | 0.66 |
ENSMUST00000114338.9
|
Add1
|
adducin 1 (alpha) |
| chr2_+_29779750 | 0.66 |
ENSMUST00000113763.8
ENSMUST00000113757.8 ENSMUST00000113756.8 ENSMUST00000133233.8 ENSMUST00000113759.9 ENSMUST00000113755.8 ENSMUST00000137558.8 ENSMUST00000046571.14 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_-_120520954 | 0.66 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr11_+_53410552 | 0.65 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr2_+_84670956 | 0.65 |
ENSMUST00000111625.2
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr6_-_86646118 | 0.65 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr1_-_177624017 | 0.64 |
ENSMUST00000016105.9
|
Adss
|
adenylosuccinate synthetase, non muscle |
| chr5_+_137756407 | 0.64 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr10_+_80972368 | 0.64 |
ENSMUST00000119606.8
ENSMUST00000146895.2 ENSMUST00000121840.8 |
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr18_-_35795233 | 0.64 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr17_+_47906985 | 0.63 |
ENSMUST00000182539.8
|
Ccnd3
|
cyclin D3 |
| chr7_-_103463120 | 0.63 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr7_+_44117444 | 0.62 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr7_-_48530777 | 0.62 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr5_+_34731087 | 0.61 |
ENSMUST00000147574.8
ENSMUST00000146295.8 |
Add1
|
adducin 1 (alpha) |
| chr6_-_71417607 | 0.61 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chrX_+_12454031 | 0.60 |
ENSMUST00000033313.3
|
Atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr2_-_104647041 | 0.59 |
ENSMUST00000117237.2
ENSMUST00000231375.2 |
Qser1
|
glutamine and serine rich 1 |
| chr5_-_138169476 | 0.56 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_44117475 | 0.55 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr15_-_101833160 | 0.54 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr7_+_44117404 | 0.54 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr17_-_71617945 | 0.53 |
ENSMUST00000232777.2
ENSMUST00000024849.11 |
Emilin2
|
elastin microfibril interfacer 2 |
| chr5_-_138169509 | 0.53 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_44117511 | 0.53 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr16_+_36832119 | 0.51 |
ENSMUST00000071452.12
ENSMUST00000054034.7 |
Polq
|
polymerase (DNA directed), theta |
| chrX_+_10583629 | 0.51 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr6_-_122587005 | 0.51 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr6_+_87755046 | 0.50 |
ENSMUST00000032133.5
|
Gp9
|
glycoprotein 9 (platelet) |
| chr4_+_123798625 | 0.50 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr2_+_126850613 | 0.50 |
ENSMUST00000110394.8
ENSMUST00000002063.15 |
Ap4e1
|
adaptor-related protein complex AP-4, epsilon 1 |
| chr5_-_138169253 | 0.50 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr3_+_14545751 | 0.49 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr6_+_116241146 | 0.48 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr11_-_34724458 | 0.48 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr18_-_67857594 | 0.48 |
ENSMUST00000120934.8
ENSMUST00000025420.14 ENSMUST00000122412.2 |
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr15_-_98851423 | 0.48 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr8_+_70953766 | 0.47 |
ENSMUST00000127983.2
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr10_+_80641067 | 0.47 |
ENSMUST00000036016.6
|
Amh
|
anti-Mullerian hormone |
| chr17_+_84013575 | 0.46 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr17_-_71617968 | 0.46 |
ENSMUST00000233057.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr3_+_63203235 | 0.46 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr5_-_37874461 | 0.46 |
ENSMUST00000094836.6
|
Stk32b
|
serine/threonine kinase 32B |
| chr6_-_51446752 | 0.45 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr9_+_62754252 | 0.45 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr13_+_81931642 | 0.44 |
ENSMUST00000224574.2
|
Cetn3
|
centrin 3 |
| chr2_+_30306045 | 0.44 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr7_-_126398343 | 0.44 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr15_-_64886753 | 0.43 |
ENSMUST00000110100.3
|
Gm21961
|
predicted gene, 21961 |
| chr6_+_52691204 | 0.43 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chrX_+_36390430 | 0.43 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr4_+_123798690 | 0.42 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr11_+_87628356 | 0.42 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chrX_-_139857424 | 0.42 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr9_-_32255556 | 0.42 |
ENSMUST00000214223.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr9_-_88364593 | 0.42 |
ENSMUST00000173801.8
ENSMUST00000069221.12 ENSMUST00000172508.2 |
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_-_146479323 | 0.42 |
ENSMUST00000032427.15
|
Ints13
|
integrator complex subunit 13 |
| chr7_-_132454332 | 0.41 |
ENSMUST00000120425.8
ENSMUST00000033257.15 |
Eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr10_-_127358231 | 0.41 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_-_78642480 | 0.41 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr9_+_107464841 | 0.41 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr7_+_90091937 | 0.41 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr15_-_79976016 | 0.41 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr5_-_110927803 | 0.40 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr1_-_120197979 | 0.40 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr11_+_72889889 | 0.40 |
ENSMUST00000021141.14
|
P2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr12_+_26519203 | 0.40 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr9_-_32255533 | 0.40 |
ENSMUST00000216033.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr11_+_60428788 | 0.40 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr1_+_134333506 | 0.39 |
ENSMUST00000027726.14
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr7_-_110581652 | 0.39 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr6_-_124790029 | 0.39 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chr1_-_52271455 | 0.38 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr10_-_127358300 | 0.38 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_126399208 | 0.38 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr10_-_116385007 | 0.37 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr9_-_45923908 | 0.37 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr5_-_107873883 | 0.37 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr15_+_78294154 | 0.37 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr9_-_103357564 | 0.37 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr10_-_30679289 | 0.37 |
ENSMUST00000215725.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_-_155848917 | 0.37 |
ENSMUST00000138762.8
|
Cep350
|
centrosomal protein 350 |
| chr11_+_96679976 | 0.36 |
ENSMUST00000093943.10
|
Cbx1
|
chromobox 1 |
| chr11_-_113600838 | 0.35 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr9_-_123507937 | 0.35 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr9_+_55056648 | 0.35 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr7_+_113113061 | 0.35 |
ENSMUST00000129087.8
ENSMUST00000067929.15 ENSMUST00000164745.8 ENSMUST00000136158.8 |
Far1
|
fatty acyl CoA reductase 1 |
| chr15_+_79975520 | 0.34 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr12_+_111387023 | 0.34 |
ENSMUST00000220852.2
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr7_-_103778992 | 0.34 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr19_+_36325683 | 0.33 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr9_-_123507847 | 0.33 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr2_-_20973337 | 0.33 |
ENSMUST00000141298.9
ENSMUST00000125783.3 |
Arhgap21
|
Rho GTPase activating protein 21 |
| chr7_-_37806912 | 0.33 |
ENSMUST00000108023.10
|
Ccne1
|
cyclin E1 |
| chr1_-_13444249 | 0.33 |
ENSMUST00000068304.13
ENSMUST00000006037.13 |
Ncoa2
|
nuclear receptor coactivator 2 |
| chr1_+_181952302 | 0.33 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr17_+_23879448 | 0.32 |
ENSMUST00000062967.10
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr14_+_54713557 | 0.32 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr19_+_5928649 | 0.32 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr5_-_92583078 | 0.31 |
ENSMUST00000038514.15
|
Nup54
|
nucleoporin 54 |
| chr6_+_146479357 | 0.31 |
ENSMUST00000067404.13
ENSMUST00000111663.9 ENSMUST00000058245.5 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr4_-_152216322 | 0.31 |
ENSMUST00000105653.8
|
Espn
|
espin |
| chrX_-_74460168 | 0.30 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr11_+_106265645 | 0.30 |
ENSMUST00000106816.8
|
Prr29
|
proline rich 29 |
| chr11_+_96680061 | 0.30 |
ENSMUST00000079702.4
|
Cbx1
|
chromobox 1 |
| chr3_-_132389584 | 0.30 |
ENSMUST00000196206.5
ENSMUST00000029663.11 |
Aimp1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr19_-_5711650 | 0.29 |
ENSMUST00000236006.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr17_+_29899420 | 0.29 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chrX_+_72108393 | 0.29 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr7_+_18618605 | 0.28 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr7_+_113113037 | 0.28 |
ENSMUST00000033018.15
|
Far1
|
fatty acyl CoA reductase 1 |
| chr7_-_126391657 | 0.28 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr5_-_66308421 | 0.28 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr3_-_144425819 | 0.27 |
ENSMUST00000199531.5
ENSMUST00000199854.5 |
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr5_-_123859070 | 0.27 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr8_+_106587268 | 0.27 |
ENSMUST00000212610.2
ENSMUST00000212484.2 ENSMUST00000212200.2 |
Nutf2
|
nuclear transport factor 2 |
| chr5_+_37332834 | 0.27 |
ENSMUST00000208827.2
ENSMUST00000207619.2 |
Gm1043
|
predicted gene 1043 |
| chr11_-_102207486 | 0.26 |
ENSMUST00000146896.9
ENSMUST00000079589.11 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr8_-_19000947 | 0.26 |
ENSMUST00000055503.12
ENSMUST00000095438.10 |
Xkr5
|
X-linked Kx blood group related 5 |
| chr11_+_96679944 | 0.26 |
ENSMUST00000018810.10
|
Cbx1
|
chromobox 1 |
| chr2_-_103627334 | 0.26 |
ENSMUST00000111147.8
|
Caprin1
|
cell cycle associated protein 1 |
| chr2_+_119505543 | 0.26 |
ENSMUST00000028767.9
|
Rtf1
|
RTF1, Paf1/RNA polymerase II complex component |
| chr1_+_129201081 | 0.25 |
ENSMUST00000073527.13
ENSMUST00000040311.14 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr6_-_143045844 | 0.25 |
ENSMUST00000204140.2
|
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr11_+_106265660 | 0.25 |
ENSMUST00000188561.7
ENSMUST00000190795.7 ENSMUST00000185986.7 ENSMUST00000190268.2 |
Prr29
|
proline rich 29 |
| chr7_+_45199259 | 0.25 |
ENSMUST00000210797.2
|
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr12_+_69771761 | 0.25 |
ENSMUST00000222950.2
|
Dmac2l
|
distal membrane arm assembly complex 2 like |
| chr10_-_127031578 | 0.25 |
ENSMUST00000038217.14
ENSMUST00000130855.8 ENSMUST00000116229.2 ENSMUST00000144322.8 |
Dtx3
|
deltex 3, E3 ubiquitin ligase |
| chrX_+_72030945 | 0.25 |
ENSMUST00000164800.8
ENSMUST00000114546.9 |
Zfp185
|
zinc finger protein 185 |
| chr11_-_98040377 | 0.25 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr5_-_145077048 | 0.25 |
ENSMUST00000031627.9
|
Pdap1
|
PDGFA associated protein 1 |
| chr16_-_32630847 | 0.25 |
ENSMUST00000179384.3
|
Smbd1
|
somatomedin B domain containing 1 |
| chr2_-_153286361 | 0.25 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr5_+_92285748 | 0.24 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr2_+_84867554 | 0.24 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr12_+_83678987 | 0.24 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chr5_-_123859153 | 0.24 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr5_+_115417725 | 0.24 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr8_+_106587212 | 0.24 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr9_+_21746785 | 0.24 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr1_+_75483721 | 0.23 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr2_+_53082079 | 0.23 |
ENSMUST00000028336.7
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr18_-_46874611 | 0.23 |
ENSMUST00000035648.6
|
Atg12
|
autophagy related 12 |
| chr16_+_32151056 | 0.23 |
ENSMUST00000115151.5
ENSMUST00000232137.2 |
Ubxn7
|
UBX domain protein 7 |
| chr1_-_84817000 | 0.23 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr4_+_3940747 | 0.23 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr18_-_35795175 | 0.23 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr12_-_33142858 | 0.23 |
ENSMUST00000095774.3
|
Cdhr3
|
cadherin-related family member 3 |
| chr7_-_43906802 | 0.23 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr3_-_40856935 | 0.22 |
ENSMUST00000099123.5
|
1700034I23Rik
|
RIKEN cDNA 1700034I23 gene |
| chr10_-_49664839 | 0.22 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr13_-_58550290 | 0.22 |
ENSMUST00000043269.14
ENSMUST00000177060.8 ENSMUST00000224182.2 ENSMUST00000176207.8 |
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr16_+_20536545 | 0.22 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_-_102207516 | 0.22 |
ENSMUST00000107115.8
ENSMUST00000128016.2 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr7_-_100306160 | 0.22 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr12_+_3622379 | 0.21 |
ENSMUST00000173199.8
ENSMUST00000164578.9 ENSMUST00000174479.8 ENSMUST00000173240.8 ENSMUST00000174663.8 ENSMUST00000173736.8 |
Dtnb
|
dystrobrevin, beta |
| chr11_+_66969119 | 0.21 |
ENSMUST00000108689.8
ENSMUST00000007301.14 ENSMUST00000165221.2 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chr14_-_34096574 | 0.21 |
ENSMUST00000023826.5
|
Sncg
|
synuclein, gamma |
| chr2_-_102903680 | 0.21 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr13_-_63579497 | 0.21 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr12_-_70158348 | 0.20 |
ENSMUST00000220689.2
|
Nin
|
ninein |
| chr5_-_66308666 | 0.20 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr13_+_51799268 | 0.20 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_+_156300325 | 0.20 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr12_-_69771604 | 0.20 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr18_+_37453427 | 0.20 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
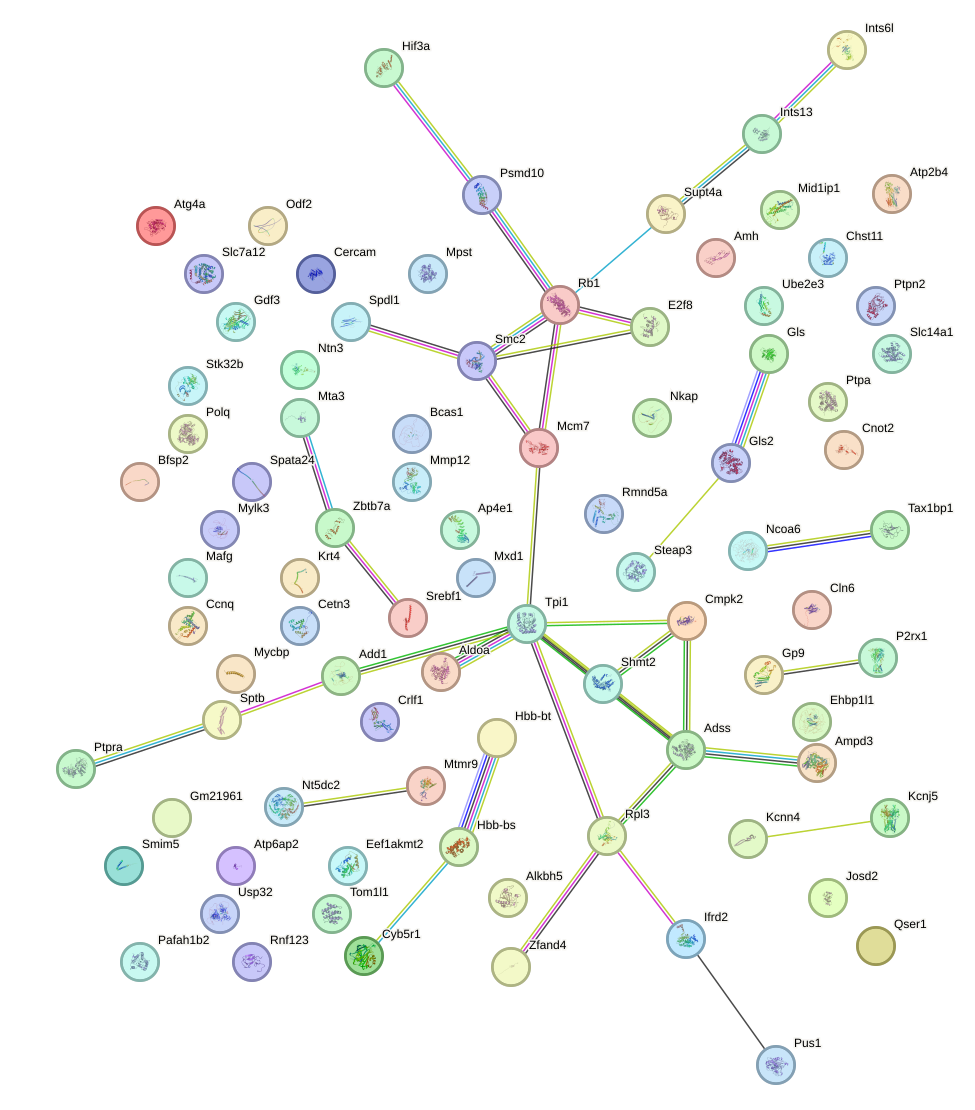
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 0.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 0.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 0.9 | GO:1903943 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 1.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.9 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 1.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.5 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 2.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.4 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.4 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.1 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.5 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.6 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 2.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.6 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.3 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.3 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.9 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 | 0.6 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.4 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.2 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.3 | GO:0045900 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.4 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:1901080 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.5 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 1.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.0 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.2 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.2 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.6 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.3 | 2.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 0.8 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 1.7 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 0.6 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 0.6 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.1 | 0.4 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.2 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 1.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 1.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 3.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 3.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |