Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zic4
Z-value: 0.97
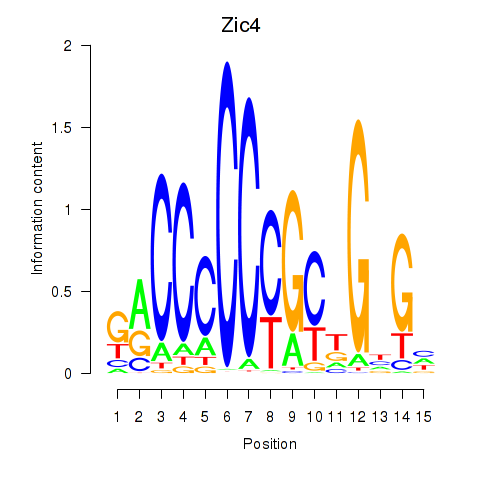
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic4
|
ENSMUSG00000036972.15 | zinc finger protein of the cerebellum 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | mm39_v1_chr9_+_91260688_91260700 | -0.23 | 1.8e-01 | Click! |
Activity profile of Zic4 motif
Sorted Z-values of Zic4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_107893676 | 4.22 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr7_-_34914675 | 2.92 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr12_-_102671154 | 2.62 |
ENSMUST00000178697.2
ENSMUST00000046518.12 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chr17_-_45904540 | 2.18 |
ENSMUST00000163905.8
ENSMUST00000167692.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_+_98932062 | 2.05 |
ENSMUST00000017637.13
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr17_-_34250616 | 1.99 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr16_-_18245352 | 1.92 |
ENSMUST00000000335.12
|
Comt
|
catechol-O-methyltransferase |
| chr11_+_101358990 | 1.87 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr17_-_34250474 | 1.84 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr15_+_7159038 | 1.77 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr11_+_57692399 | 1.77 |
ENSMUST00000020826.6
|
Sap30l
|
SAP30-like |
| chr4_-_124744454 | 1.66 |
ENSMUST00000125776.8
ENSMUST00000163946.2 ENSMUST00000106190.10 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr1_+_131890679 | 1.65 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr7_+_28466160 | 1.65 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr19_+_4905158 | 1.64 |
ENSMUST00000119694.3
ENSMUST00000237504.2 ENSMUST00000237011.2 |
Ctsf
|
cathepsin F |
| chr9_-_107109108 | 1.55 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr4_+_141095415 | 1.54 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr17_+_26332260 | 1.53 |
ENSMUST00000235821.2
ENSMUST00000025010.14 ENSMUST00000237058.2 |
Pgap6
|
post-glycosylphosphatidylinositol attachment to proteins 6 |
| chr4_+_152423344 | 1.52 |
ENSMUST00000005175.5
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr7_+_142660971 | 1.48 |
ENSMUST00000009689.11
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr17_-_45906768 | 1.47 |
ENSMUST00000164618.8
ENSMUST00000097317.10 ENSMUST00000170113.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_-_124744266 | 1.39 |
ENSMUST00000137769.3
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr13_-_95359543 | 1.38 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr7_+_127400016 | 1.34 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_+_127399776 | 1.30 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr14_-_32186721 | 1.29 |
ENSMUST00000191501.2
|
Slc18a3
|
solute carrier family 18 (vesicular monoamine), member 3 |
| chr12_-_98541293 | 1.25 |
ENSMUST00000110113.3
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr14_-_47426863 | 1.21 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr12_-_98540944 | 1.20 |
ENSMUST00000221305.2
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr7_-_97228589 | 1.20 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr1_-_13061333 | 1.18 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chrX_+_137815171 | 1.18 |
ENSMUST00000064937.14
ENSMUST00000113052.8 |
Nrk
|
Nik related kinase |
| chr16_+_20551853 | 1.16 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr6_+_83031502 | 1.16 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr7_-_44320244 | 1.14 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr9_-_58462720 | 1.12 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr7_+_27290969 | 1.11 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_+_27291126 | 1.11 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_+_127399789 | 1.10 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_+_115373895 | 1.09 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr11_+_98932586 | 1.08 |
ENSMUST00000177092.8
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr2_-_143853122 | 1.05 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr12_-_5425682 | 1.05 |
ENSMUST00000020958.9
|
Klhl29
|
kelch-like 29 |
| chr6_-_115228800 | 1.04 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr10_+_77522403 | 1.01 |
ENSMUST00000092366.4
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr2_-_152793376 | 0.98 |
ENSMUST00000123121.9
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr14_-_24054352 | 0.97 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_-_29323500 | 0.95 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr17_+_37356872 | 0.95 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_127399848 | 0.95 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr17_-_24752683 | 0.95 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr4_+_152423075 | 0.95 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr14_+_33662976 | 0.95 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr14_-_118289557 | 0.95 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr13_+_15637831 | 0.94 |
ENSMUST00000141194.8
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr4_+_124744472 | 0.93 |
ENSMUST00000102628.11
|
Yrdc
|
yrdC domain containing (E.coli) |
| chr10_-_81186025 | 0.90 |
ENSMUST00000122993.8
|
Hmg20b
|
high mobility group 20B |
| chr2_-_152793469 | 0.90 |
ENSMUST00000037715.7
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr2_-_25124240 | 0.90 |
ENSMUST00000006638.8
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chrX_+_41238410 | 0.89 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
| chr15_+_74435587 | 0.89 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr19_+_47167444 | 0.88 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr17_-_29456750 | 0.88 |
ENSMUST00000137727.3
|
Cpne5
|
copine V |
| chr14_+_70768289 | 0.86 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr11_+_99755302 | 0.86 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr11_-_78313043 | 0.85 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr14_+_70768257 | 0.85 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr6_+_22288220 | 0.84 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr11_-_102837514 | 0.84 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr2_-_152793607 | 0.83 |
ENSMUST00000109811.4
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr17_-_83939316 | 0.83 |
ENSMUST00000234613.2
ENSMUST00000051482.2 |
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr7_-_16020668 | 0.82 |
ENSMUST00000150528.9
ENSMUST00000118976.9 ENSMUST00000146609.3 |
Ccdc9
|
coiled-coil domain containing 9 |
| chr2_-_165076609 | 0.81 |
ENSMUST00000065438.13
|
Cdh22
|
cadherin 22 |
| chr19_+_48194464 | 0.81 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr7_+_49624978 | 0.81 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr19_-_60456742 | 0.80 |
ENSMUST00000051277.4
|
Prlhr
|
prolactin releasing hormone receptor |
| chr8_+_93687561 | 0.80 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr6_-_39095144 | 0.79 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr2_-_144173615 | 0.78 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr10_-_81186137 | 0.78 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr6_-_48422759 | 0.77 |
ENSMUST00000114561.9
|
Zfp467
|
zinc finger protein 467 |
| chr4_-_129015027 | 0.76 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr1_-_166066298 | 0.76 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr17_-_51486196 | 0.76 |
ENSMUST00000024717.10
ENSMUST00000224528.2 |
Tbc1d5
|
TBC1 domain family, member 5 |
| chr15_-_97902576 | 0.75 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr11_-_78388284 | 0.74 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr2_+_120807498 | 0.73 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr1_-_37758863 | 0.73 |
ENSMUST00000160589.2
|
Cracdl
|
capping protein inhibiting regulator of actin like |
| chr13_+_15637786 | 0.72 |
ENSMUST00000130065.8
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr19_+_37423198 | 0.72 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr11_+_69891398 | 0.69 |
ENSMUST00000019362.15
ENSMUST00000190940.2 |
Dvl2
|
dishevelled segment polarity protein 2 |
| chr7_+_139414057 | 0.69 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr11_-_116226175 | 0.69 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chr13_+_15638466 | 0.69 |
ENSMUST00000110510.4
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr15_-_97902515 | 0.67 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr8_-_45835234 | 0.67 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr11_-_59181573 | 0.66 |
ENSMUST00000010044.8
|
Wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr14_-_24054186 | 0.66 |
ENSMUST00000188991.7
ENSMUST00000224468.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr13_+_34221572 | 0.66 |
ENSMUST00000040656.8
|
Bphl
|
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
| chr6_+_99669640 | 0.65 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr5_+_139529643 | 0.65 |
ENSMUST00000174792.2
|
Uncx
|
UNC homeobox |
| chr17_+_25936463 | 0.65 |
ENSMUST00000115108.4
|
Gng13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr14_-_24054273 | 0.62 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr13_-_95359955 | 0.62 |
ENSMUST00000159608.8
|
Pde8b
|
phosphodiesterase 8B |
| chr2_-_160754212 | 0.61 |
ENSMUST00000109454.8
ENSMUST00000057169.5 |
Emilin3
|
elastin microfibril interfacer 3 |
| chr12_-_75224099 | 0.60 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr11_-_55498559 | 0.59 |
ENSMUST00000108853.8
ENSMUST00000075603.5 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr8_+_123844090 | 0.59 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chr15_+_99122742 | 0.57 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr11_-_101062111 | 0.57 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr4_+_62278932 | 0.56 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr19_+_16416664 | 0.55 |
ENSMUST00000237350.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr11_+_108812474 | 0.55 |
ENSMUST00000144511.2
|
Axin2
|
axin 2 |
| chr14_-_24053994 | 0.55 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_+_141470261 | 0.54 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr11_-_78388560 | 0.54 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr5_-_113458563 | 0.54 |
ENSMUST00000154248.8
ENSMUST00000112325.8 ENSMUST00000048112.13 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr15_+_75781584 | 0.53 |
ENSMUST00000192937.2
|
Tigd5
|
tigger transposable element derived 5 |
| chr14_+_119375753 | 0.53 |
ENSMUST00000065904.5
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr19_+_5497575 | 0.53 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr4_+_125384481 | 0.53 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr4_-_108075119 | 0.52 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr3_-_63836796 | 0.52 |
ENSMUST00000061706.7
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr3_+_54063459 | 0.51 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr6_+_61157279 | 0.50 |
ENSMUST00000126214.8
|
Ccser1
|
coiled-coil serine rich 1 |
| chr4_-_24851086 | 0.49 |
ENSMUST00000084781.6
ENSMUST00000108218.10 |
Klhl32
|
kelch-like 32 |
| chr2_-_166904902 | 0.49 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr13_-_63721412 | 0.49 |
ENSMUST00000195106.2
|
Ptch1
|
patched 1 |
| chrX_-_110606766 | 0.48 |
ENSMUST00000113422.9
ENSMUST00000038472.7 |
Hdx
|
highly divergent homeobox |
| chr6_+_141470105 | 0.48 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr3_+_133015846 | 0.48 |
ENSMUST00000029644.16
ENSMUST00000122334.8 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr7_+_49624612 | 0.47 |
ENSMUST00000151721.8
ENSMUST00000081872.13 |
Nell1
|
NEL-like 1 |
| chr7_+_28466658 | 0.47 |
ENSMUST00000155327.8
|
Sirt2
|
sirtuin 2 |
| chr11_+_80191692 | 0.47 |
ENSMUST00000017836.8
|
Rhbdl3
|
rhomboid like 3 |
| chr10_+_7465555 | 0.46 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr4_+_42917228 | 0.46 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr2_+_164721277 | 0.46 |
ENSMUST00000041643.5
|
Pcif1
|
phosphorylated CTD interacting factor 1 |
| chr12_-_112893382 | 0.46 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr8_-_106863521 | 0.45 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_-_4527475 | 0.45 |
ENSMUST00000059295.10
|
Syt12
|
synaptotagmin XII |
| chr10_+_45211847 | 0.45 |
ENSMUST00000095715.5
|
Bves
|
blood vessel epicardial substance |
| chr7_+_45522551 | 0.43 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr17_+_35055962 | 0.43 |
ENSMUST00000173874.8
ENSMUST00000180043.8 ENSMUST00000046244.15 |
Dxo
|
decapping exoribonuclease |
| chr1_+_153541020 | 0.43 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_129015493 | 0.42 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr5_-_125418107 | 0.42 |
ENSMUST00000111390.8
ENSMUST00000086075.13 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr9_+_87026337 | 0.42 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr17_-_85995680 | 0.42 |
ENSMUST00000024947.8
ENSMUST00000163568.4 |
Six2
|
sine oculis-related homeobox 2 |
| chr5_-_49682150 | 0.40 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr6_-_126621751 | 0.40 |
ENSMUST00000055168.5
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr6_-_126621770 | 0.40 |
ENSMUST00000203094.2
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr13_+_55097200 | 0.40 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr19_-_6285827 | 0.40 |
ENSMUST00000025695.10
|
Ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr11_-_70128462 | 0.39 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr15_+_61858883 | 0.39 |
ENSMUST00000159338.2
|
Myc
|
myelocytomatosis oncogene |
| chr1_-_194859015 | 0.39 |
ENSMUST00000082321.9
ENSMUST00000195120.6 |
Cr2
|
complement receptor 2 |
| chr12_-_67269323 | 0.38 |
ENSMUST00000037181.16
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr15_-_64794139 | 0.38 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr5_-_76452365 | 0.38 |
ENSMUST00000075159.5
|
Clock
|
circadian locomotor output cycles kaput |
| chr10_+_7465845 | 0.38 |
ENSMUST00000135907.8
|
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr9_-_105372330 | 0.38 |
ENSMUST00000038118.15
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr17_+_86475205 | 0.38 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr19_+_6144449 | 0.37 |
ENSMUST00000235513.2
|
Gm550
|
predicted gene 550 |
| chr17_+_37356854 | 0.37 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_+_92990110 | 0.37 |
ENSMUST00000107863.4
|
Car10
|
carbonic anhydrase 10 |
| chr19_+_47167554 | 0.37 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr7_+_6442835 | 0.36 |
ENSMUST00000168341.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr7_-_12514534 | 0.36 |
ENSMUST00000211392.2
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr14_+_26414422 | 0.36 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr2_-_180929828 | 0.35 |
ENSMUST00000049032.13
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr10_+_58649181 | 0.35 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr5_-_66308421 | 0.33 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr8_-_33374282 | 0.33 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr7_-_67409432 | 0.33 |
ENSMUST00000208815.2
ENSMUST00000074233.12 ENSMUST00000208231.2 ENSMUST00000051389.10 |
Synm
|
synemin, intermediate filament protein |
| chr8_-_88686188 | 0.32 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr17_+_37357451 | 0.32 |
ENSMUST00000172789.2
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr5_-_49682106 | 0.32 |
ENSMUST00000176191.8
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr16_+_90628001 | 0.32 |
ENSMUST00000099543.10
|
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr11_-_55499014 | 0.32 |
ENSMUST00000102716.10
|
Glra1
|
glycine receptor, alpha 1 subunit |
| chr7_-_132725575 | 0.31 |
ENSMUST00000171968.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_-_119910986 | 0.31 |
ENSMUST00000134319.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr8_-_106863423 | 0.31 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_+_97229065 | 0.30 |
ENSMUST00000107153.3
|
Rsf1
|
remodeling and spacing factor 1 |
| chr4_+_116953218 | 0.30 |
ENSMUST00000030443.12
|
Ptch2
|
patched 2 |
| chr2_+_164802729 | 0.29 |
ENSMUST00000202623.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr2_-_144174066 | 0.29 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr10_-_79473675 | 0.29 |
ENSMUST00000020564.7
|
Shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr7_+_143838191 | 0.28 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_186437760 | 0.28 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr14_-_24054927 | 0.28 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_+_93768175 | 0.27 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr1_-_194858918 | 0.27 |
ENSMUST00000210219.2
ENSMUST00000193801.2 |
Cr2
|
complement receptor 2 |
| chr5_-_142495408 | 0.26 |
ENSMUST00000110784.8
|
Radil
|
Ras association and DIL domains |
| chr5_+_4242367 | 0.25 |
ENSMUST00000177258.2
|
Mterf1b
|
mitochondrial transcription termination factor 1b |
| chr8_-_112120442 | 0.25 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr4_+_42158092 | 0.24 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr5_+_115573055 | 0.24 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr7_-_46783432 | 0.24 |
ENSMUST00000102626.10
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr1_+_153541412 | 0.24 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_-_58247764 | 0.24 |
ENSMUST00000112608.9
ENSMUST00000112607.3 ENSMUST00000028178.14 |
Acvr1c
|
activin A receptor, type IC |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.7 | 2.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.7 | 4.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 3.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 3.1 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.5 | 1.5 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.5 | 1.9 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.4 | 4.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.4 | 1.6 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 1.2 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.4 | 1.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.4 | 2.2 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.3 | 0.9 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 2.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 1.2 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.3 | 1.8 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 2.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.2 | 0.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.7 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.7 | GO:0061184 | COP9 signalosome assembly(GO:0010387) negative regulation of cardioblast differentiation(GO:0051892) positive regulation of dermatome development(GO:0061184) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 | 0.8 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.2 | 1.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 3.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.2 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 0.8 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 0.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.2 | 0.5 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 2.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.9 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 2.6 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 1.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.7 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.1 | 1.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 1.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 1.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.4 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.4 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 2.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.3 | GO:0070237 | endocardial cushion fusion(GO:0003274) positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.1 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.3 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 0.5 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 2.1 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.8 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 1.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.4 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.9 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.7 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.0 | 0.8 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.3 | 1.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 1.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 3.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.8 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.2 | 0.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 1.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.8 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 2.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.9 | 2.6 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.8 | 0.8 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.6 | 1.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.6 | 2.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.5 | 2.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.4 | 1.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.4 | 1.6 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 3.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 1.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 1.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.3 | 0.9 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 3.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 1.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.7 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 1.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 1.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.9 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 0.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 4.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.6 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 0.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 2.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 2.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.4 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 1.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0035276 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 2.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |