Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
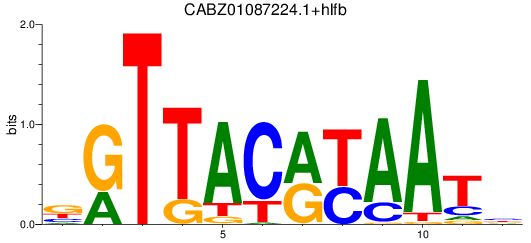
Results for CABZ01087224.1+hlfb
Z-value: 0.90
Transcription factors associated with CABZ01087224.1+hlfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlfb
|
ENSDARG00000061011 | HLF transcription factor, PAR bZIP family member b |
|
CABZ01087224.1
|
ENSDARG00000111269 | ENSDARG00000111269 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hlfb | dr11_v1_chr12_+_32159272_32159272 | -0.20 | 4.8e-02 | Click! |
| CABZ01087224.1 | dr11_v1_chr3_+_11568523_11568523 | 0.05 | 6.1e-01 | Click! |
Activity profile of CABZ01087224.1+hlfb motif
Sorted Z-values of CABZ01087224.1+hlfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50147948 | 14.51 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr1_-_10071422 | 14.28 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr2_+_10134345 | 13.15 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr5_+_45677781 | 12.15 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr9_-_16109001 | 11.85 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr3_-_40275096 | 11.77 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr16_-_45917322 | 10.92 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr9_-_48736388 | 8.90 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_-_63509581 | 8.71 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr20_-_40755614 | 8.28 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr14_+_16345003 | 7.84 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr21_-_27443995 | 7.17 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr13_-_25196758 | 6.92 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr22_+_15315655 | 6.88 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr25_-_13188678 | 6.87 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr16_-_45917683 | 6.53 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr14_+_3507326 | 6.41 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr15_-_1885247 | 6.40 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr25_-_29415369 | 6.25 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr20_-_40750953 | 6.16 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr13_-_36034582 | 5.96 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr22_+_10606863 | 5.95 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr22_+_10606573 | 5.85 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr13_+_10945337 | 5.83 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr12_+_48390715 | 5.74 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr6_+_9130989 | 5.67 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr19_-_34970312 | 5.57 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr15_-_29348212 | 5.40 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr13_-_10945288 | 5.34 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr7_+_56577522 | 5.19 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr25_+_13191391 | 5.17 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr21_+_45841731 | 5.10 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr7_+_12950507 | 5.09 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr12_-_20584413 | 5.04 |
ENSDART00000170923
|
FP885542.2
|
|
| chr20_-_25522911 | 4.96 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr7_+_67733759 | 4.95 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr14_+_32430982 | 4.86 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr7_+_38380135 | 4.76 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr5_+_29831235 | 4.73 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr25_+_13191615 | 4.70 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr1_+_14253118 | 4.53 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr5_-_2721686 | 4.28 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr21_+_17051478 | 4.24 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_-_12725513 | 4.24 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr9_+_38292947 | 4.18 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr5_+_25585869 | 4.16 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr7_+_15736230 | 4.15 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr24_-_32665283 | 4.13 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr18_-_16922905 | 4.12 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr4_-_27099224 | 4.05 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr17_-_15149192 | 4.02 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr22_+_15336752 | 4.02 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr17_-_6076266 | 3.98 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr6_+_42475730 | 3.94 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr22_-_17677947 | 3.87 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr11_+_25504215 | 3.81 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr6_-_26895750 | 3.78 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr8_-_21103041 | 3.73 |
ENSDART00000171771
ENSDART00000131322 ENSDART00000137838 |
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr9_+_1654284 | 3.71 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr14_+_4151379 | 3.70 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr18_+_25546227 | 3.69 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr6_+_10333920 | 3.68 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr24_-_26328721 | 3.64 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr11_-_23501467 | 3.62 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr5_-_20185665 | 3.62 |
ENSDART00000051612
|
dao.2
|
D-amino-acid oxidase, tandem duplicate 2 |
| chr11_-_23459779 | 3.55 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr12_-_20373058 | 3.54 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr22_+_24645325 | 3.50 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr20_-_7293837 | 3.44 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr24_+_38671054 | 3.41 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr19_+_14113886 | 3.41 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr5_-_69212184 | 3.38 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr3_+_30921246 | 3.34 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr11_-_42750626 | 3.30 |
ENSDART00000130640
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr20_-_25533739 | 3.28 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr20_+_35857399 | 3.25 |
ENSDART00000102611
|
cd2ap
|
CD2-associated protein |
| chr8_-_21103522 | 3.22 |
ENSDART00000100283
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr18_-_25645680 | 3.21 |
ENSDART00000135874
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr2_+_22851832 | 3.20 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr2_-_51772438 | 3.17 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr11_-_45138857 | 3.11 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr17_-_43031763 | 3.10 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr22_+_11756040 | 3.10 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr15_+_19838458 | 3.08 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr25_-_13202208 | 3.02 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr18_-_7539166 | 2.92 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr19_+_43715911 | 2.87 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr16_+_32029090 | 2.86 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr23_-_36441693 | 2.86 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr12_+_33396489 | 2.86 |
ENSDART00000149960
|
fasn
|
fatty acid synthase |
| chr17_-_6076084 | 2.84 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr15_-_17960228 | 2.83 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr12_+_19199735 | 2.83 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr17_+_25833947 | 2.83 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr25_+_32473433 | 2.81 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr22_+_11775269 | 2.80 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr3_-_26183699 | 2.80 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr19_+_5674907 | 2.79 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr5_-_2112030 | 2.78 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr17_-_10838434 | 2.71 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr20_+_25563105 | 2.70 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr7_-_12968689 | 2.69 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr15_+_26600611 | 2.68 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr3_+_7763114 | 2.67 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr13_-_25774183 | 2.67 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr16_-_29387215 | 2.67 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr11_+_43419809 | 2.59 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr2_-_37277626 | 2.58 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr12_-_22400999 | 2.57 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr10_-_20523405 | 2.55 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr8_-_25846188 | 2.55 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr17_+_51744450 | 2.54 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr4_+_5531583 | 2.54 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr19_-_12965020 | 2.54 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr16_-_19568388 | 2.51 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr22_+_997838 | 2.51 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr24_-_26310854 | 2.51 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr17_-_20236228 | 2.50 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr5_-_33259079 | 2.48 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr2_+_52232630 | 2.47 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr6_+_23122789 | 2.46 |
ENSDART00000049226
ENSDART00000067560 |
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr19_+_14109348 | 2.46 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr15_+_12435975 | 2.39 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr3_+_1107102 | 2.38 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr6_-_39649504 | 2.35 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr15_-_24178893 | 2.33 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr12_-_1557286 | 2.29 |
ENSDART00000082110
|
sstr2a
|
somatostatin receptor 2a |
| chr6_+_50451337 | 2.29 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr7_-_46019756 | 2.28 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr5_+_13870340 | 2.27 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr25_+_32473277 | 2.24 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr7_+_34297271 | 2.24 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr24_-_36727922 | 2.22 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr1_-_354115 | 2.22 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr19_+_11985572 | 2.21 |
ENSDART00000130537
|
spag1a
|
sperm associated antigen 1a |
| chr15_-_1765098 | 2.21 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr14_+_8174828 | 2.20 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr9_-_21838045 | 2.19 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr4_+_9478500 | 2.18 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr17_+_26965351 | 2.18 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr12_-_23365737 | 2.12 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr10_-_15053507 | 2.12 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr14_-_38929885 | 2.11 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr19_+_16015881 | 2.10 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr7_-_38792543 | 2.09 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr24_+_32525146 | 2.09 |
ENSDART00000132417
ENSDART00000110185 |
yme1l1a
|
YME1-like 1a |
| chr22_+_15973122 | 2.08 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr7_+_38090515 | 2.07 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr20_+_2039518 | 2.07 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr4_+_21866851 | 2.07 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr4_+_5341592 | 2.06 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr7_-_10560964 | 2.06 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr9_-_9225980 | 2.05 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr4_-_25796848 | 2.05 |
ENSDART00000122881
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr2_-_55779927 | 2.04 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr3_-_57744323 | 2.04 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr2_-_19234329 | 2.03 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr24_-_26854032 | 2.02 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr5_+_27421639 | 2.02 |
ENSDART00000146285
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr14_+_1170968 | 2.01 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr21_+_1647990 | 2.00 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr21_+_29179887 | 1.98 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr25_-_12803723 | 1.98 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr13_+_25397098 | 1.97 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr3_+_36972298 | 1.96 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr5_+_26138313 | 1.94 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr13_+_25396896 | 1.94 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr20_-_30900947 | 1.94 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr11_+_39107131 | 1.94 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr6_-_39080630 | 1.93 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr9_+_30478768 | 1.91 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr24_-_23784701 | 1.90 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr17_+_17764979 | 1.89 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr9_+_41218967 | 1.88 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr1_-_23595779 | 1.88 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr20_-_33909872 | 1.87 |
ENSDART00000153018
|
atf6
|
activating transcription factor 6 |
| chr15_-_35960250 | 1.87 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr10_+_23099890 | 1.85 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr6_-_1553314 | 1.82 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr21_+_43178831 | 1.81 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr24_-_17049270 | 1.80 |
ENSDART00000175508
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr3_-_32927516 | 1.80 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr16_-_21785261 | 1.80 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr23_-_25686894 | 1.78 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr17_+_23578316 | 1.77 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr10_-_23099809 | 1.77 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr25_+_32474031 | 1.76 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr25_-_26833100 | 1.76 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr20_-_9760424 | 1.75 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr10_-_2875735 | 1.73 |
ENSDART00000034555
|
ddx56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr16_-_33104944 | 1.72 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr5_-_11809404 | 1.71 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr22_-_10121880 | 1.70 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_-_24757231 | 1.70 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr23_+_20640875 | 1.69 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr19_+_16016038 | 1.67 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr20_+_38201644 | 1.66 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr22_+_26443235 | 1.66 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr5_+_56277866 | 1.66 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr12_-_9132682 | 1.65 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01087224.1+hlfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 2.3 | 6.8 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.9 | 5.7 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 1.7 | 5.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.5 | 4.5 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 1.5 | 6.0 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.4 | 5.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.4 | 9.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.4 | 5.4 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 1.2 | 11.8 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 1.2 | 3.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 1.2 | 6.9 | GO:0044209 | AMP salvage(GO:0044209) |
| 1.0 | 4.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.0 | 4.0 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 1.0 | 3.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.0 | 14.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.8 | 2.5 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.8 | 5.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.8 | 14.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.7 | 2.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.7 | 3.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.7 | 2.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.7 | 2.0 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.7 | 3.4 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 3.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.6 | 3.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.6 | 2.3 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.6 | 2.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.6 | 18.0 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.6 | 3.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.6 | 2.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.6 | 1.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.5 | 2.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.5 | 16.9 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.5 | 2.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.5 | 4.9 | GO:0010885 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.5 | 3.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 2.8 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.5 | 3.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 12.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.4 | 3.1 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.4 | 2.6 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.4 | 1.2 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.4 | 2.8 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.4 | 10.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 1.9 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.4 | 3.7 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.4 | 2.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.1 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 1.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 5.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 2.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.3 | 4.7 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.3 | 4.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.3 | 4.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.3 | 2.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.3 | 2.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.3 | 1.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.3 | 2.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 1.9 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.7 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 3.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.2 | 2.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 20.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.2 | 2.4 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 1.0 | GO:1902804 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.8 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 6.1 | GO:0007568 | aging(GO:0007568) |
| 0.2 | 0.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 0.9 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.9 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 0.7 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.9 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 0.5 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 2.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.2 | 14.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 2.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 0.8 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.6 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.3 | GO:0006560 | proline metabolic process(GO:0006560) proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 1.4 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 4.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.7 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 1.5 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 1.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.3 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.6 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 5.9 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 | 0.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.6 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 1.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 1.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 1.1 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 1.4 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 4.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 2.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 3.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.9 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 2.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 3.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.2 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.2 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.1 | 1.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 2.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 7.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.4 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 3.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.9 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 2.8 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 1.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 3.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.6 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 3.9 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.2 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.5 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.1 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.4 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 1.9 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 1.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 2.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 2.9 | GO:0044272 | sulfur compound biosynthetic process(GO:0044272) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 4.9 | GO:0009725 | response to hormone(GO:0009725) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 1.3 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 2.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 2.8 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.7 | GO:0030041 | actin filament polymerization(GO:0030041) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.4 | 4.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.8 | 3.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 1.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 2.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.5 | 3.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.4 | 1.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.3 | 5.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 1.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.3 | 1.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 1.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 3.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 0.9 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 4.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 14.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 17.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.8 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 3.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 2.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 5.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 4.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 10.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 71.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 9.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 8.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 5.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 3.9 | 11.8 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 2.5 | 7.5 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 2.3 | 6.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 2.3 | 6.8 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 1.6 | 6.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 1.4 | 5.7 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.4 | 4.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.4 | 6.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.1 | 4.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.1 | 8.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.0 | 4.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 1.0 | 2.9 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.8 | 2.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 3.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.8 | 1.6 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.8 | 3.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.7 | 2.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 5.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.5 | 2.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.5 | 2.7 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.5 | 2.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.5 | 5.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 1.9 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.5 | 2.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 1.4 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.5 | 1.8 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 2.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.4 | 2.0 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.4 | 3.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 2.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 2.3 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 14.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.4 | 1.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.3 | 2.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 10.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 7.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 4.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 2.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 1.2 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.3 | 2.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 0.6 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.3 | 2.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.3 | 1.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.3 | 0.8 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 3.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 4.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.0 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 1.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 2.0 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 6.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 10.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 2.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.2 | 1.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 34.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 8.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 11.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 5.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 2.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 3.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 3.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 8.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 4.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.2 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 4.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 7.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 3.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 3.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 3.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 3.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 5.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 6.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 10.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 2.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.1 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 5.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 15.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.4 | 16.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.9 | 11.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.9 | 6.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.7 | 6.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.7 | 9.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 6.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 12.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 4.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 8.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 2.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 14.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 1.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 1.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 2.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 5.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |