Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
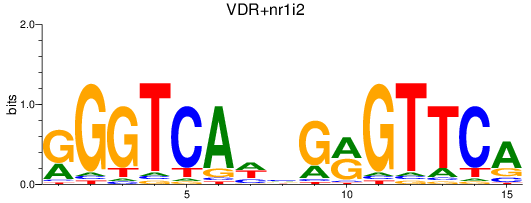
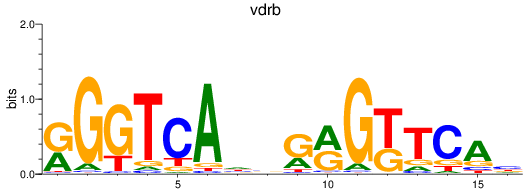
Results for VDR+nr1i2_vdrb
Z-value: 0.65
Transcription factors associated with VDR+nr1i2_vdrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1i2
|
ENSDARG00000029766 | nuclear receptor subfamily 1, group I, member 2 |
|
VDR
|
ENSDARG00000112923 | vitamin D receptor |
|
vdrb
|
ENSDARG00000070721 | vitamin D receptor b |
|
vdrb
|
ENSDARG00000114864 | vitamin D receptor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vdrb | dr11_v1_chr6_-_39005133_39005133 | -0.22 | 3.7e-02 | Click! |
| CABZ01088036.1 | dr11_v1_chr23_+_45785563_45785563 | -0.16 | 1.2e-01 | Click! |
| nr1i2 | dr11_v1_chr9_+_9502610_9502610 | -0.12 | 2.5e-01 | Click! |
Activity profile of VDR+nr1i2_vdrb motif
Sorted Z-values of VDR+nr1i2_vdrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_4148719 | 17.00 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr8_+_1769475 | 4.30 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr2_+_10134345 | 4.17 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr1_-_25679339 | 3.97 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr14_-_21063977 | 3.88 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr23_-_44723102 | 3.87 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr14_-_21064199 | 3.84 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr3_+_46425262 | 3.13 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr3_+_12744083 | 3.09 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr20_+_17581881 | 2.97 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr25_+_13191391 | 2.66 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr6_-_13498745 | 2.47 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr3_-_32912787 | 2.41 |
ENSDART00000156149
|
g6pcb
|
glucose-6-phosphatase b, catalytic subunit |
| chr21_-_25756119 | 2.32 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr7_+_52122224 | 2.23 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr6_+_9893554 | 2.16 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr14_+_22076596 | 2.14 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr25_+_13191615 | 2.13 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr21_-_1012269 | 1.97 |
ENSDART00000159835
|
CABZ01057159.1
|
|
| chr5_-_42544522 | 1.84 |
ENSDART00000157968
|
BX510949.1
|
|
| chr10_+_7564106 | 1.78 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr7_-_38183331 | 1.74 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr7_+_46003449 | 1.71 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr25_+_4541211 | 1.70 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr23_+_42454292 | 1.67 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| chr23_+_44611864 | 1.67 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr21_+_5589923 | 1.63 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr6_+_9155271 | 1.62 |
ENSDART00000168727
|
zgc:64065
|
zgc:64065 |
| chr10_+_7563755 | 1.55 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr10_-_373575 | 1.48 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr15_+_32297441 | 1.43 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr21_+_29179887 | 1.42 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr12_+_20506197 | 1.41 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr9_-_28616436 | 1.41 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr2_-_17392799 | 1.34 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr16_+_16969060 | 1.34 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr23_-_46040618 | 1.33 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr21_-_19918286 | 1.32 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr9_-_7378566 | 1.30 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr14_+_8715527 | 1.28 |
ENSDART00000170754
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr19_-_20148469 | 1.28 |
ENSDART00000134476
|
si:ch211-155k24.1
|
si:ch211-155k24.1 |
| chr14_-_41468892 | 1.24 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr10_-_22803740 | 1.23 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr2_+_21309272 | 1.20 |
ENSDART00000141322
|
zbtb47a
|
zinc finger and BTB domain containing 47a |
| chr7_-_24995631 | 1.19 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr25_-_13188214 | 1.12 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr3_+_52473670 | 1.11 |
ENSDART00000154310
|
si:ch211-191c10.3
|
si:ch211-191c10.3 |
| chr1_-_23595779 | 1.06 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr13_+_45332203 | 1.05 |
ENSDART00000110466
|
si:ch211-168h21.3
|
si:ch211-168h21.3 |
| chr12_-_1557286 | 1.05 |
ENSDART00000082110
|
sstr2a
|
somatostatin receptor 2a |
| chr14_-_11430566 | 1.04 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr24_+_2495197 | 1.01 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr19_+_2896188 | 0.98 |
ENSDART00000151421
|
si:ch211-251g8.5
|
si:ch211-251g8.5 |
| chr8_+_23245596 | 0.96 |
ENSDART00000143313
|
si:ch211-196c10.11
|
si:ch211-196c10.11 |
| chr18_+_7073130 | 0.93 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr12_+_46608361 | 0.93 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr16_+_19536614 | 0.92 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr17_-_32370047 | 0.92 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr3_+_36366577 | 0.89 |
ENSDART00000151203
|
rgs9a
|
regulator of G protein signaling 9a |
| chr4_-_11737939 | 0.88 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr3_-_34656745 | 0.87 |
ENSDART00000144824
|
znfl1
|
zinc finger-like gene 1 |
| chr12_+_18663154 | 0.84 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr20_+_34374735 | 0.84 |
ENSDART00000144090
|
si:dkeyp-11g8.3
|
si:dkeyp-11g8.3 |
| chr20_-_29505863 | 0.83 |
ENSDART00000148278
|
klf11a
|
Kruppel-like factor 11a |
| chr15_-_24883956 | 0.81 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr12_+_2660733 | 0.81 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr9_-_31763246 | 0.81 |
ENSDART00000114742
|
si:dkey-250i3.3
|
si:dkey-250i3.3 |
| chr6_+_50451337 | 0.79 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr8_+_25295160 | 0.77 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| chr14_-_11529311 | 0.72 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr3_+_32749613 | 0.70 |
ENSDART00000053684
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr9_+_15837398 | 0.68 |
ENSDART00000141063
|
si:dkey-103d23.5
|
si:dkey-103d23.5 |
| chr20_-_40760217 | 0.67 |
ENSDART00000101007
|
cx34.5
|
connexin 34.5 |
| chr6_+_27624023 | 0.65 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr4_-_18416566 | 0.65 |
ENSDART00000033717
|
cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr4_-_20177868 | 0.64 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr22_-_8720794 | 0.64 |
ENSDART00000191236
ENSDART00000185738 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr10_+_42691210 | 0.64 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr3_-_25116208 | 0.64 |
ENSDART00000183628
|
chadla
|
chondroadherin-like a |
| chr15_+_20543770 | 0.63 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr10_+_15603082 | 0.63 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr21_-_18972206 | 0.62 |
ENSDART00000146743
|
pimr72
|
Pim proto-oncogene, serine/threonine kinase, related 72 |
| chr9_+_38888025 | 0.62 |
ENSDART00000148306
|
map2
|
microtubule-associated protein 2 |
| chr15_+_9053059 | 0.61 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr7_+_3558512 | 0.61 |
ENSDART00000131884
|
si:dkey-192d15.1
|
si:dkey-192d15.1 |
| chr23_+_28809002 | 0.56 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr6_-_43677125 | 0.56 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr10_+_9561066 | 0.56 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr15_-_38361787 | 0.56 |
ENSDART00000155758
|
trpc2a
|
transient receptor potential cation channel, subfamily C, member 2a |
| chr19_-_32600638 | 0.55 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr5_+_39875646 | 0.55 |
ENSDART00000097608
|
si:ch73-138i16.1
|
si:ch73-138i16.1 |
| chr1_+_54626491 | 0.54 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr23_+_35672542 | 0.54 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr8_+_20918207 | 0.52 |
ENSDART00000144039
|
si:ch73-196i15.5
|
si:ch73-196i15.5 |
| chr23_+_5631381 | 0.50 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr8_-_3346692 | 0.50 |
ENSDART00000057874
|
FUT9 (1 of many)
|
zgc:103510 |
| chrM_+_11299 | 0.49 |
ENSDART00000093618
|
mt-nd4
|
NADH dehydrogenase 4, mitochondrial |
| chr18_+_36037223 | 0.49 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr12_-_13556536 | 0.49 |
ENSDART00000166396
|
CR354395.1
|
|
| chr14_+_35383267 | 0.48 |
ENSDART00000143818
|
clint1a
|
clathrin interactor 1a |
| chr22_-_30999489 | 0.48 |
ENSDART00000139706
|
ssuh2.3
|
ssu-2 homolog, tandem duplicate 3 |
| chr14_-_24101897 | 0.48 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr19_-_9776075 | 0.48 |
ENSDART00000140332
|
si:dkey-14o18.1
|
si:dkey-14o18.1 |
| chr14_-_858985 | 0.47 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr24_-_32563875 | 0.46 |
ENSDART00000020982
|
CR792422.1
|
|
| chr15_-_8665662 | 0.45 |
ENSDART00000090675
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr11_-_3907937 | 0.44 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr8_-_1267247 | 0.43 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr16_+_5539189 | 0.42 |
ENSDART00000178106
|
CR848788.2
|
|
| chr25_+_23591990 | 0.42 |
ENSDART00000151938
ENSDART00000089199 |
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr20_+_263056 | 0.42 |
ENSDART00000132669
|
tube1
|
tubulin, epsilon 1 |
| chr8_-_3336273 | 0.42 |
ENSDART00000081160
|
zgc:173737
|
zgc:173737 |
| chr21_+_39197418 | 0.41 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr7_+_47243564 | 0.41 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr19_-_35400819 | 0.39 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr14_+_36521005 | 0.39 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr5_+_40224938 | 0.38 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr8_+_20956078 | 0.38 |
ENSDART00000136397
|
si:dkeyp-82a1.2
|
si:dkeyp-82a1.2 |
| chr20_+_18993452 | 0.37 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr1_-_56951369 | 0.37 |
ENSDART00000152294
|
si:ch211-1f22.2
|
si:ch211-1f22.2 |
| chr22_-_7168571 | 0.37 |
ENSDART00000165549
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr10_+_23548674 | 0.37 |
ENSDART00000079686
|
zmp:0000001103
|
zmp:0000001103 |
| chr3_-_29977495 | 0.37 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr6_-_48087152 | 0.36 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_+_36366053 | 0.36 |
ENSDART00000181140
|
rgs9a
|
regulator of G protein signaling 9a |
| chr18_+_33788340 | 0.36 |
ENSDART00000136950
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr18_+_33762090 | 0.36 |
ENSDART00000147605
|
si:dkey-145c18.4
|
si:dkey-145c18.4 |
| chr4_-_30528239 | 0.35 |
ENSDART00000169290
|
znf1052
|
zinc finger protein 1052 |
| chr10_-_24319526 | 0.35 |
ENSDART00000148480
|
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr21_+_39197628 | 0.35 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr16_+_41135018 | 0.35 |
ENSDART00000139435
|
si:ch211-53m15.2
|
si:ch211-53m15.2 |
| chr12_-_30359031 | 0.34 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr12_-_25217217 | 0.33 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr12_-_30358836 | 0.33 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr1_-_40914752 | 0.33 |
ENSDART00000113087
|
hmx1
|
H6 family homeobox 1 |
| chr11_-_25324534 | 0.33 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr24_+_17345521 | 0.32 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr5_-_37252111 | 0.32 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr15_-_23699737 | 0.32 |
ENSDART00000078311
|
zgc:154093
|
zgc:154093 |
| chr4_-_29833326 | 0.31 |
ENSDART00000150596
|
si:ch211-231i17.4
|
si:ch211-231i17.4 |
| chr4_-_16345227 | 0.31 |
ENSDART00000079521
|
kera
|
keratocan |
| chr6_-_9922266 | 0.31 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr22_+_34694217 | 0.30 |
ENSDART00000111793
|
hykk.1
|
hydroxylysine kinase, tandem duplicate 1 |
| chr23_-_10831995 | 0.30 |
ENSDART00000142533
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr6_-_29049022 | 0.30 |
ENSDART00000190309
|
evi5b
|
ecotropic viral integration site 5b |
| chr4_-_49875682 | 0.29 |
ENSDART00000185921
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr21_-_31286428 | 0.29 |
ENSDART00000185334
|
ca4c
|
carbonic anhydrase IV c |
| chr14_+_36521553 | 0.29 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr13_-_31866276 | 0.28 |
ENSDART00000140164
|
syt14a
|
synaptotagmin XIVa |
| chr14_+_22297257 | 0.28 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr3_+_19336286 | 0.27 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr16_-_22747125 | 0.27 |
ENSDART00000179865
|
tlr19
|
toll-like receptor 19 |
| chr11_+_2642013 | 0.27 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr12_-_36070473 | 0.26 |
ENSDART00000165720
|
gpr142
|
G protein-coupled receptor 142 |
| chr20_+_48100261 | 0.26 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr1_-_57129179 | 0.26 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr8_-_44015210 | 0.25 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr8_-_15991801 | 0.24 |
ENSDART00000132005
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr7_-_3721671 | 0.24 |
ENSDART00000062732
|
si:ch211-282j17.8
|
si:ch211-282j17.8 |
| chr17_+_15674052 | 0.24 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr23_+_21966447 | 0.24 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr21_-_28235361 | 0.24 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr24_+_19986432 | 0.23 |
ENSDART00000184402
|
CR381686.5
|
|
| chr15_-_28805493 | 0.23 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr13_+_2448251 | 0.22 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr17_-_50071748 | 0.22 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr5_+_22459087 | 0.22 |
ENSDART00000134781
|
BX546499.1
|
|
| chr4_+_9450415 | 0.22 |
ENSDART00000189690
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr22_+_35205968 | 0.21 |
ENSDART00000150467
|
tsc22d2
|
TSC22 domain family 2 |
| chr16_+_5901835 | 0.21 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr24_+_1294176 | 0.20 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr20_+_23960525 | 0.20 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr15_+_2421729 | 0.20 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr9_-_53422084 | 0.20 |
ENSDART00000162661
|
gpc6b
|
glypican 6b |
| chr1_+_39865748 | 0.19 |
ENSDART00000131954
ENSDART00000130270 |
irf2a
|
interferon regulatory factor 2a |
| chr2_-_22575851 | 0.19 |
ENSDART00000182932
|
FQ377628.2
|
|
| chr20_+_18992679 | 0.19 |
ENSDART00000147105
ENSDART00000152811 |
tdh
|
L-threonine dehydrogenase |
| chr10_-_40964739 | 0.19 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr21_+_25216397 | 0.18 |
ENSDART00000130970
|
sycn.3
|
syncollin, tandem duplicate 3 |
| chr15_-_25198107 | 0.18 |
ENSDART00000159793
|
mnta
|
MAX network transcriptional repressor a |
| chr17_+_38602602 | 0.18 |
ENSDART00000187996
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr19_+_37701450 | 0.17 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr3_-_41795917 | 0.17 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr3_+_17939828 | 0.17 |
ENSDART00000185047
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr2_+_25278107 | 0.17 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr14_-_25309058 | 0.16 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr16_+_23600041 | 0.16 |
ENSDART00000186918
|
kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr25_+_17860962 | 0.15 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr1_-_56223913 | 0.15 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr6_-_39080630 | 0.14 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr23_+_1276006 | 0.14 |
ENSDART00000162294
|
utrn
|
utrophin |
| chr22_-_4880603 | 0.14 |
ENSDART00000134055
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr24_-_18446604 | 0.12 |
ENSDART00000144203
|
sec61g
|
Sec61 translocon gamma subunit |
| chr25_+_27923846 | 0.12 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr9_-_6208067 | 0.12 |
ENSDART00000032368
|
tmtops2b
|
teleost multiple tissue opsin 2b |
| chr9_-_10778615 | 0.12 |
ENSDART00000182577
|
CABZ01053619.1
|
|
| chr15_+_20530649 | 0.11 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr12_+_48581588 | 0.11 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr19_-_32600138 | 0.11 |
ENSDART00000052101
|
zgc:91944
|
zgc:91944 |
| chr5_-_68927728 | 0.11 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR+nr1i2_vdrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 1.0 | 3.0 | GO:0090247 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.4 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 1.7 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 5.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 17.0 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.3 | 0.9 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.9 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.0 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.5 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 1.0 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 3.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 8.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.6 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 3.5 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 2.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.7 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.1 | 0.6 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.4 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 2.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.5 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.3 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.2 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 5.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.0 | 1.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 3.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.6 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 18.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 4.2 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 3.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 1.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.2 | 1.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.6 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 4.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 6.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.7 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0019840 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 3.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 13.2 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.7 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 17.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 4.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 3.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |