Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
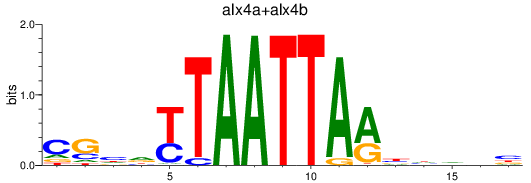
Results for alx4a+alx4b
Z-value: 0.52
Transcription factors associated with alx4a+alx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx4b
|
ENSDARG00000074442 | ALX homeobox 4b |
|
alx4a
|
ENSDARG00000088332 | ALX homeobox 4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx4a | dr11_v1_chr7_-_26924903_26924903 | 0.20 | 5.8e-02 | Click! |
| alx4b | dr11_v1_chr18_+_38321039_38321039 | -0.13 | 1.9e-01 | Click! |
Activity profile of alx4a+alx4b motif
Sorted Z-values of alx4a+alx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_31713239 | 3.99 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr13_-_20381485 | 3.81 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr5_+_2815021 | 3.67 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr12_-_35830625 | 3.65 |
ENSDART00000180028
|
CU459056.1
|
|
| chr15_-_5815006 | 3.40 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr9_+_34127005 | 3.06 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr19_-_5699703 | 2.94 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr6_-_607063 | 2.94 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr14_+_32430982 | 2.77 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr22_-_23748284 | 2.73 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr20_+_53441935 | 2.69 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr7_+_6652967 | 2.64 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr15_-_462783 | 2.59 |
ENSDART00000161049
|
CABZ01033205.1
|
Danio rerio uncharacterized LOC100002960 (LOC100002960), mRNA. |
| chr1_+_127250 | 2.54 |
ENSDART00000003463
|
f7i
|
coagulation factor VIIi |
| chr25_-_13569234 | 2.35 |
ENSDART00000189645
|
fa2h
|
fatty acid 2-hydroxylase |
| chr21_-_19314618 | 2.31 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr21_+_45841731 | 2.25 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr10_+_21867307 | 2.19 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr2_-_17392799 | 2.19 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr22_+_19552987 | 2.15 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr4_-_9891874 | 1.95 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr8_-_30979494 | 1.87 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr20_-_33566640 | 1.87 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr10_+_11261576 | 1.86 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr10_+_26747755 | 1.85 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr7_-_71531846 | 1.85 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr8_+_24745041 | 1.76 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr6_-_58764672 | 1.58 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr18_-_25568994 | 1.56 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr3_+_3681116 | 1.56 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr18_+_7073130 | 1.56 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr2_-_17393216 | 1.55 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr17_+_51627209 | 1.49 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr14_-_4145594 | 1.48 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr13_+_646700 | 1.48 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr13_-_22862133 | 1.47 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr20_-_23439011 | 1.46 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr24_-_32582378 | 1.44 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr1_-_7951002 | 1.43 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr12_-_4243268 | 1.43 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr16_+_40563533 | 1.40 |
ENSDART00000190368
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr8_+_43340995 | 1.39 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr13_-_50565338 | 1.32 |
ENSDART00000062684
|
blnk
|
B cell linker |
| chr17_-_37395460 | 1.32 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr15_-_47848544 | 1.29 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr24_-_40860603 | 1.28 |
ENSDART00000188032
|
CU633479.7
|
|
| chr15_-_25527580 | 1.27 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr10_+_6013076 | 1.27 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr6_+_612594 | 1.27 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr6_-_7720332 | 1.26 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr17_-_49412313 | 1.23 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr11_-_669558 | 1.22 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr10_+_17776981 | 1.21 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr24_-_32582880 | 1.19 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr10_-_13343831 | 1.19 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr23_+_4709607 | 1.17 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr15_-_21014270 | 1.16 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr21_+_25236297 | 1.14 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr19_-_5669122 | 1.14 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr17_-_2039511 | 1.13 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr3_-_3798755 | 1.12 |
ENSDART00000170535
|
gucy2c
|
guanylate cyclase 2C |
| chr11_-_45138857 | 1.12 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_31609481 | 1.11 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr25_+_10811551 | 1.10 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr8_-_12867128 | 1.10 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr7_-_66877058 | 1.09 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr14_-_5817039 | 1.09 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr15_-_1885247 | 1.08 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr11_-_45141309 | 1.08 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr4_+_77971104 | 1.07 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr9_+_28598577 | 1.03 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr19_+_19988869 | 1.03 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr16_-_28658341 | 1.03 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_32054159 | 1.03 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr2_-_24407933 | 1.03 |
ENSDART00000088584
|
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr4_-_1908179 | 1.02 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr13_-_31008275 | 1.02 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr6_-_43677125 | 1.01 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr4_+_25692277 | 1.00 |
ENSDART00000047113
|
acot18
|
acyl-CoA thioesterase 18 |
| chr18_-_5527050 | 0.99 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr24_-_25166720 | 0.99 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_-_29197700 | 0.98 |
ENSDART00000172614
|
si:ch211-57b15.2
|
si:ch211-57b15.2 |
| chr2_+_25315591 | 0.97 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr12_+_20641471 | 0.97 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr12_+_47081783 | 0.93 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr3_-_49925313 | 0.93 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr10_-_31015535 | 0.91 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr23_+_26733232 | 0.91 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr14_-_14659023 | 0.90 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr16_-_22544047 | 0.90 |
ENSDART00000131657
|
cgna
|
cingulin a |
| chr18_-_34549721 | 0.90 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr1_+_52560549 | 0.90 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr12_-_18578218 | 0.88 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr18_+_22109379 | 0.88 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr24_-_11076400 | 0.87 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr4_+_16715267 | 0.86 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr25_-_21031007 | 0.86 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_+_39695827 | 0.86 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr15_+_32398921 | 0.85 |
ENSDART00000156125
ENSDART00000153806 |
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr18_+_14684115 | 0.83 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr8_-_12867434 | 0.83 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr7_-_66868543 | 0.83 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr22_-_10541372 | 0.83 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr7_-_8712148 | 0.82 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr2_-_57110477 | 0.81 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr8_+_23738122 | 0.81 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr18_+_2228737 | 0.81 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr12_+_20641102 | 0.81 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr18_+_14619544 | 0.80 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr16_-_16761164 | 0.80 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr15_+_5360407 | 0.79 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr23_-_33350990 | 0.79 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr21_+_39941875 | 0.78 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr11_+_42474694 | 0.78 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr4_+_18824959 | 0.78 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr12_+_22580579 | 0.77 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr15_-_2519640 | 0.76 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr20_-_9095105 | 0.75 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr5_+_71802014 | 0.75 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr15_+_32798333 | 0.75 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr12_+_5358409 | 0.75 |
ENSDART00000152632
|
plce1
|
phospholipase C, epsilon 1 |
| chr24_+_5912635 | 0.74 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr24_-_5911973 | 0.74 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr10_-_42882215 | 0.72 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr23_-_20051369 | 0.72 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr1_+_55755304 | 0.72 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr7_+_57088920 | 0.71 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr22_-_10156581 | 0.71 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr3_+_29941777 | 0.71 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr21_+_21195487 | 0.70 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr16_+_23303859 | 0.69 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr22_+_7738966 | 0.69 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr14_+_33329420 | 0.69 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr10_+_41199660 | 0.66 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr8_+_25034544 | 0.65 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr24_+_21621654 | 0.65 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr8_+_31435452 | 0.65 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr12_+_37401331 | 0.64 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr3_-_29941357 | 0.64 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr7_-_34448076 | 0.64 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_-_6085750 | 0.64 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr5_+_66312359 | 0.64 |
ENSDART00000137534
|
malt1
|
MALT paracaspase 1 |
| chr1_-_49250490 | 0.64 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr17_-_27048537 | 0.64 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr3_-_34136368 | 0.63 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr8_-_31062811 | 0.63 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr25_+_20715950 | 0.63 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr20_-_49889111 | 0.63 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr8_-_45838277 | 0.63 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr15_-_3736773 | 0.63 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr10_+_35358675 | 0.62 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr3_-_61494840 | 0.62 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr8_-_8698607 | 0.62 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr17_+_24821627 | 0.62 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr20_+_48116476 | 0.61 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr20_+_36812368 | 0.61 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr13_-_36798204 | 0.61 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr8_+_28695914 | 0.61 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr9_-_38587275 | 0.60 |
ENSDART00000077446
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr2_-_28396993 | 0.60 |
ENSDART00000188170
|
CABZ01056052.1
|
|
| chr15_-_19051152 | 0.60 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr12_-_18578432 | 0.60 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr16_+_50953842 | 0.60 |
ENSDART00000174709
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr8_-_45760087 | 0.59 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr9_-_53666031 | 0.59 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr20_-_43663494 | 0.58 |
ENSDART00000144564
|
BX470188.1
|
|
| chr13_+_18321140 | 0.58 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr16_-_38118003 | 0.58 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr5_+_27898226 | 0.58 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr16_+_50953547 | 0.57 |
ENSDART00000157526
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr8_-_21091961 | 0.57 |
ENSDART00000100281
|
aldh9a1a.2
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 2 |
| chr18_+_41527877 | 0.57 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr19_-_27858033 | 0.57 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr13_-_12602920 | 0.56 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr22_-_10541712 | 0.56 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr21_-_25801956 | 0.55 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr6_-_39270851 | 0.55 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr7_-_8738827 | 0.55 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr12_+_47698356 | 0.55 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr2_-_56348727 | 0.55 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr23_-_33361425 | 0.55 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr6_+_36821621 | 0.55 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr11_+_44617021 | 0.54 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr13_+_7442023 | 0.54 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr20_+_51197758 | 0.54 |
ENSDART00000020084
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr25_-_37191929 | 0.54 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr18_+_8320165 | 0.54 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr6_-_55399214 | 0.53 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr19_+_43780970 | 0.53 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr6_+_9427641 | 0.53 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_+_2169337 | 0.53 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr12_-_28363111 | 0.52 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr8_-_25034411 | 0.52 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr6_+_3717613 | 0.51 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr15_+_31820536 | 0.51 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr16_+_42471455 | 0.51 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr8_+_53064920 | 0.51 |
ENSDART00000164823
|
nadka
|
NAD kinase a |
| chr15_+_28318005 | 0.51 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr4_+_77943184 | 0.50 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_4532066 | 0.50 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx4a+alx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 2.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 1.4 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.4 | 1.3 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.4 | 1.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.4 | 1.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.4 | 2.5 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 1.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 2.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.9 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 0.6 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.2 | 1.1 | GO:0070178 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 1.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 0.5 | GO:0071634 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 | 1.6 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 0.5 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 0.5 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 0.5 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 10.6 | GO:0007599 | hemostasis(GO:0007599) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 1.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.4 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 2.4 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.1 | 0.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.9 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.4 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.2 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.8 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 2.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.6 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.9 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0044857 | membrane raft assembly(GO:0001765) membrane raft organization(GO:0031579) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 1.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 2.9 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.3 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 3.1 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.9 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 1.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.9 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 1.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.5 | GO:0072350 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.7 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.6 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.2 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0072584 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.4 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.5 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.3 | 1.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 17.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 15.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.8 | 2.3 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.5 | 3.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.4 | 1.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.4 | 1.6 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.3 | 1.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 1.0 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 2.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.3 | 0.9 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 2.7 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.3 | 2.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 1.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 0.8 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 1.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.9 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 1.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.9 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 0.9 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 0.7 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 1.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 2.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.3 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 1.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.3 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 0.3 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.4 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 12.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 2.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.7 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0038187 | pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |