Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
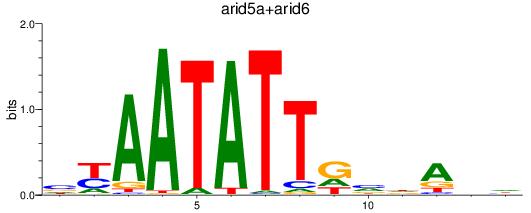
Results for arid5a+arid6
Z-value: 0.86
Transcription factors associated with arid5a+arid6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
|
arid5a
|
ENSDARG00000077120 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid5a | dr11_v1_chr8_+_52377516_52377516 | 0.15 | 1.6e-01 | Click! |
| arid6 | dr11_v1_chr21_+_11244068_11244068 | 0.08 | 4.7e-01 | Click! |
Activity profile of arid5a+arid6 motif
Sorted Z-values of arid5a+arid6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_3585934 | 7.29 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr7_-_12968689 | 7.24 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr15_-_21155641 | 6.44 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr7_+_39385540 | 5.95 |
ENSDART00000139240
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr7_+_10610791 | 5.60 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr1_+_10051763 | 5.41 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr15_-_452347 | 5.12 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr20_+_46925581 | 5.12 |
ENSDART00000192531
|
si:ch73-21k16.5
|
si:ch73-21k16.5 |
| chr7_+_39446247 | 4.97 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr22_-_24880824 | 4.91 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr13_-_9895564 | 4.87 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr23_+_26017227 | 4.81 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_+_36457888 | 4.73 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr4_-_8043839 | 4.70 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr15_-_21165237 | 4.68 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr15_-_26552393 | 4.56 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr17_+_43867889 | 4.52 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr1_+_29068654 | 4.49 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr1_-_59313465 | 4.41 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr16_+_26777473 | 4.41 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr14_-_1454045 | 4.31 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr22_+_25753972 | 4.30 |
ENSDART00000188417
|
AL929192.2
|
|
| chr22_-_24858042 | 4.30 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr21_-_7940043 | 4.26 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr5_+_32221755 | 4.18 |
ENSDART00000125917
|
myhc4
|
myosin heavy chain 4 |
| chr7_-_30926030 | 4.14 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr3_-_61205711 | 4.08 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr22_+_25693295 | 4.04 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr25_+_7494181 | 4.03 |
ENSDART00000165005
|
cat
|
catalase |
| chr19_+_9295244 | 4.00 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr7_-_30925798 | 3.97 |
ENSDART00000149303
|
sord
|
sorbitol dehydrogenase |
| chr14_+_51056605 | 3.88 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr15_-_26552652 | 3.78 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr8_+_1766206 | 3.78 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr3_-_50139860 | 3.77 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr24_-_11446156 | 3.68 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr24_-_17039638 | 3.67 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr13_-_12021566 | 3.67 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr11_-_34097418 | 3.60 |
ENSDART00000028301
ENSDART00000138579 |
tdo2b
|
tryptophan 2,3-dioxygenase b |
| chr22_+_25687525 | 3.57 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25704430 | 3.55 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr19_+_43119014 | 3.55 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr22_+_25681911 | 3.51 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_1170294 | 3.48 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr3_+_31039923 | 3.40 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr22_-_557965 | 3.39 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr12_+_47663419 | 3.37 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr3_+_12816362 | 3.36 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr19_+_14113886 | 3.32 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr16_+_28754403 | 3.29 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr5_+_28797771 | 3.28 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr14_+_11458044 | 3.26 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr4_+_391297 | 3.24 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr22_+_25715925 | 3.23 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr12_-_17863467 | 3.16 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr22_-_17611742 | 3.11 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr14_+_11457500 | 3.11 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr4_+_74928675 | 3.09 |
ENSDART00000192755
|
nup50
|
nucleoporin 50 |
| chr16_-_9694822 | 3.04 |
ENSDART00000168748
|
COLEC10
|
si:ch211-93i7.4 |
| chr20_-_5267600 | 3.00 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr7_+_20645121 | 2.99 |
ENSDART00000052922
|
sat2a
|
spermidine/spermine N1-acetyltransferase family member 2a |
| chr18_-_19456269 | 2.98 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr5_-_38820046 | 2.97 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr22_+_19552987 | 2.95 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr2_-_37960688 | 2.92 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr20_+_54024559 | 2.91 |
ENSDART00000130767
|
si:dkey-241l7.4
|
si:dkey-241l7.4 |
| chr16_-_45917683 | 2.79 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr16_-_48400639 | 2.75 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr25_-_29074064 | 2.74 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr13_+_46941930 | 2.66 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr6_+_7322587 | 2.60 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr24_-_9991153 | 2.47 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr6_+_7466223 | 2.46 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr9_-_34191627 | 2.46 |
ENSDART00000142664
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr5_-_2721686 | 2.45 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr3_+_39566999 | 2.44 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr24_-_26328721 | 2.44 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr25_+_23280220 | 2.40 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr3_-_5228841 | 2.39 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr22_+_21317597 | 2.37 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr11_-_44906703 | 2.32 |
ENSDART00000180591
|
zgc:171772
|
zgc:171772 |
| chr24_-_25166416 | 2.25 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_-_57110477 | 2.23 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr22_+_31039091 | 2.20 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr21_-_5077715 | 2.19 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr25_+_31323978 | 2.16 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr7_+_30626378 | 2.12 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr23_+_383782 | 2.11 |
ENSDART00000055148
|
zgc:101663
|
zgc:101663 |
| chr5_-_69621227 | 2.11 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr24_+_26328787 | 2.11 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr17_+_18031304 | 2.02 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr24_-_32582378 | 2.01 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr2_+_21048661 | 1.98 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr16_+_29492749 | 1.97 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr6_+_3680651 | 1.96 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr15_-_23647078 | 1.96 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr9_-_42696408 | 1.92 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr17_-_4395373 | 1.90 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr25_-_774350 | 1.89 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr19_-_5769553 | 1.81 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr21_+_974555 | 1.80 |
ENSDART00000130648
|
scarb2c
|
scavenger receptor class B, member 2c |
| chr14_+_164556 | 1.80 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr15_+_44201056 | 1.79 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr21_-_22689805 | 1.79 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr6_+_29086594 | 1.79 |
ENSDART00000127695
|
rpl5b
|
ribosomal protein L5b |
| chr13_-_349952 | 1.77 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr22_-_2959005 | 1.77 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr1_+_32054159 | 1.76 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr12_+_46404307 | 1.76 |
ENSDART00000185011
|
BX005305.4
|
|
| chr19_+_30884960 | 1.75 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr12_+_20583552 | 1.75 |
ENSDART00000170035
|
arsg
|
arylsulfatase G |
| chr23_-_18609475 | 1.73 |
ENSDART00000104524
|
zgc:158296
|
zgc:158296 |
| chr7_+_8361083 | 1.71 |
ENSDART00000102091
|
jac7
|
jacalin 7 |
| chr3_+_57820913 | 1.71 |
ENSDART00000168101
|
CU571328.1
|
|
| chr10_-_39130839 | 1.71 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr3_+_3641429 | 1.70 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr12_+_13091842 | 1.70 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr6_-_8466717 | 1.67 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr12_+_46483618 | 1.66 |
ENSDART00000186970
|
BX005305.5
|
|
| chr21_+_10577527 | 1.66 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr5_-_42059869 | 1.66 |
ENSDART00000193984
|
cenpv
|
centromere protein V |
| chr1_-_53714885 | 1.66 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr24_-_31306724 | 1.65 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr6_-_42003780 | 1.65 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr3_-_36824190 | 1.64 |
ENSDART00000172943
|
abcc6b.2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 2 |
| chr21_+_20903244 | 1.64 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr7_+_30178253 | 1.64 |
ENSDART00000002075
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr10_-_9190831 | 1.64 |
ENSDART00000169301
|
si:dkeyp-41f9.4
|
si:dkeyp-41f9.4 |
| chr13_-_30996072 | 1.61 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr4_+_17671164 | 1.61 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr22_+_737211 | 1.60 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr23_-_45407631 | 1.59 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr23_+_642395 | 1.59 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr21_-_4849029 | 1.59 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr13_-_28689717 | 1.58 |
ENSDART00000101582
|
pcgf6
|
polycomb group ring finger 6 |
| chr11_+_44135351 | 1.58 |
ENSDART00000182914
|
FO704721.1
|
|
| chr25_+_6049639 | 1.58 |
ENSDART00000128816
|
sv2
|
synaptic vesicle glycoprotein 2 |
| chr16_-_46660680 | 1.57 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr25_+_3318192 | 1.57 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr4_-_1914228 | 1.56 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr20_+_14789305 | 1.55 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr12_-_47648538 | 1.54 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr23_+_20431388 | 1.54 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr6_+_27338512 | 1.54 |
ENSDART00000155004
|
klhl30
|
kelch-like family member 30 |
| chr15_-_1765098 | 1.54 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr7_-_30177691 | 1.54 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr24_-_25691020 | 1.52 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr17_-_31579715 | 1.51 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr12_-_979789 | 1.50 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr7_+_20467549 | 1.49 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr17_+_6585333 | 1.49 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr19_-_8798178 | 1.49 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr12_+_46443477 | 1.48 |
ENSDART00000191873
|
BX005305.1
|
|
| chr1_+_53714734 | 1.47 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr10_-_17222083 | 1.46 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr7_+_30178453 | 1.46 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr12_-_10567188 | 1.46 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr11_-_3535537 | 1.45 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr21_+_43702016 | 1.44 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr12_+_45238292 | 1.44 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr25_+_3099073 | 1.43 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr18_-_19484919 | 1.42 |
ENSDART00000177690
ENSDART00000184949 |
lctlb
|
lactase-like b |
| chr12_+_46462090 | 1.40 |
ENSDART00000130748
|
BX005305.2
|
|
| chr10_+_15024772 | 1.40 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr9_+_32178050 | 1.39 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr12_+_46386983 | 1.39 |
ENSDART00000183982
|
BX005305.3
|
Danio rerio legumain (LOC100005356), mRNA. |
| chr8_+_54135642 | 1.39 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr7_+_6879534 | 1.38 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr7_+_21275152 | 1.36 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr5_-_72058646 | 1.35 |
ENSDART00000112955
|
PRRC2B
|
proline rich coiled-coil 2B |
| chr23_+_19655301 | 1.34 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr7_-_31759394 | 1.34 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr12_+_20700961 | 1.34 |
ENSDART00000016099
|
si:ch211-119c20.2
|
si:ch211-119c20.2 |
| chr15_+_31820536 | 1.33 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr2_-_32262287 | 1.33 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr23_+_28381260 | 1.33 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr25_-_2355107 | 1.33 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr7_+_39399747 | 1.31 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr10_+_44699734 | 1.31 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr4_+_1761029 | 1.29 |
ENSDART00000026963
|
pmpcb
|
peptidase (mitochondrial processing) beta |
| chr11_+_21076872 | 1.28 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr21_-_8003782 | 1.27 |
ENSDART00000126792
|
si:dkey-163m14.7
|
si:dkey-163m14.7 |
| chr24_-_35561672 | 1.27 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr19_+_4076259 | 1.27 |
ENSDART00000160633
|
zgc:173581
|
zgc:173581 |
| chr3_+_24207243 | 1.27 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr12_+_46425800 | 1.27 |
ENSDART00000191965
|
BX005305.1
|
|
| chr11_+_37905630 | 1.26 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr14_-_4120636 | 1.25 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr4_+_13909398 | 1.25 |
ENSDART00000187959
ENSDART00000184926 |
pphln1
|
periphilin 1 |
| chr5_-_33886495 | 1.24 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr19_-_10295173 | 1.24 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr12_+_48220584 | 1.23 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr2_-_57834983 | 1.23 |
ENSDART00000185970
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr11_+_25504215 | 1.23 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr1_+_11822 | 1.22 |
ENSDART00000166393
|
cep97
|
centrosomal protein 97 |
| chr17_-_15546862 | 1.20 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr15_-_15968883 | 1.20 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr1_+_158034 | 1.19 |
ENSDART00000160843
|
cul4a
|
cullin 4A |
| chr2_+_19195841 | 1.19 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr20_+_18993452 | 1.18 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5a+arid6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 3.4 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 1.0 | 8.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 1.0 | 4.9 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.9 | 5.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.9 | 4.5 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.8 | 4.9 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.7 | 2.7 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.7 | 3.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 4.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.5 | 1.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.5 | 1.6 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.5 | 1.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 3.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.4 | 2.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.4 | 1.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.4 | 2.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 1.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.4 | 1.5 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.4 | 1.5 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.4 | 5.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 1.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 3.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.3 | 2.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 3.4 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.3 | 1.0 | GO:0032602 | chemokine production(GO:0032602) |
| 0.3 | 0.9 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.3 | 3.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 4.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 0.9 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.3 | 1.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 3.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 0.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 4.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 1.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 3.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 2.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 1.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 1.7 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.2 | 1.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.9 | GO:0006297 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.9 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.2 | 1.3 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 17.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 5.0 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.2 | 1.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.9 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 11.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 1.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 2.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 0.8 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.6 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 2.0 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 1.2 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 1.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 2.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.9 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 3.7 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 2.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 4.7 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.6 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.8 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.0 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 2.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.9 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 1.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 4.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 1.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 2.3 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 2.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 3.5 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.1 | 1.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.4 | GO:2000319 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T cell differentiation(GO:0045581) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of alpha-beta T cell differentiation(GO:0046639) regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 2.4 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.6 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.1 | 1.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 2.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 4.0 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.7 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.3 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.4 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.2 | GO:0090184 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 1.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.8 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.8 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.8 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 2.2 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.5 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 5.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 1.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 2.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.7 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 2.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 4.2 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 1.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 3.2 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.5 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.2 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 9.7 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.0 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 5.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 3.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 6.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 2.3 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.1 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.9 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.6 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.9 | GO:0010324 | membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.0 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.8 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 4.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.8 | 2.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 3.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 2.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 2.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 0.8 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 1.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 3.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 0.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.8 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 4.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 0.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 4.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 10.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 6.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 2.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.4 | GO:1990077 | primosome complex(GO:1990077) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.3 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.5 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 13.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.2 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 3.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.2 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 4.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 5.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 24.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.2 | 3.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.1 | 5.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.0 | 7.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.9 | 4.5 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.9 | 2.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.7 | 2.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.7 | 2.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.6 | 3.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 4.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 3.0 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.5 | 2.5 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.4 | 4.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 2.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.4 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 4.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.4 | 1.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 2.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 4.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 1.2 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 1.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 1.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 2.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.9 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 0.9 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.2 | 0.8 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 3.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.7 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 3.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 6.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 3.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 2.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 2.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 2.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 17.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.8 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 21.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.0 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.9 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.6 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.1 | 1.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 4.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.3 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 1.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.9 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.6 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 2.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 7.8 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.1 | 1.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 15.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.9 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 3.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 3.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 2.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 2.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 3.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 3.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 3.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.6 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 1.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 3.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 5.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 3.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 1.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 2.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 4.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 5.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 1.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 4.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 10.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF DNA | Genes involved in Synthesis of DNA |
| 0.0 | 1.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 4.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |