Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
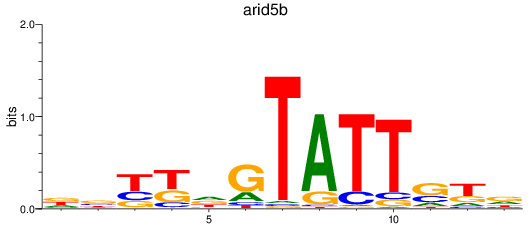
Results for arid5b
Z-value: 0.51
Transcription factors associated with arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid5b
|
ENSDARG00000037196 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid5b | dr11_v1_chr12_+_8373525_8373525 | -0.45 | 4.7e-06 | Click! |
Activity profile of arid5b motif
Sorted Z-values of arid5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_19299309 | 7.90 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr1_+_10051763 | 6.46 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr11_+_13224281 | 5.93 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr21_-_1625976 | 5.19 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr17_-_2584423 | 3.85 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2578026 | 3.80 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_60831082 | 3.40 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr13_-_4704353 | 2.99 |
ENSDART00000171900
|
oit3
|
oncoprotein induced transcript 3 |
| chr13_-_6252498 | 2.89 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr21_-_2322102 | 2.76 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr3_-_61205711 | 2.63 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr4_-_20043484 | 2.55 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr9_+_5862040 | 2.46 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr1_+_9994811 | 2.41 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr25_-_32363341 | 2.41 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr15_-_5624361 | 2.24 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr16_-_25606235 | 2.18 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr2_-_58142854 | 2.07 |
ENSDART00000169909
|
CABZ01110881.1
|
|
| chr2_-_17114852 | 1.86 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr23_-_27589754 | 1.83 |
ENSDART00000138381
ENSDART00000133721 |
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr9_-_2892045 | 1.79 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr9_-_2892250 | 1.75 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr16_+_27224068 | 1.74 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr12_+_31783066 | 1.74 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr19_-_10330778 | 1.68 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr16_-_50175069 | 1.58 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr2_-_37744951 | 1.39 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr18_-_16953978 | 1.35 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr6_-_49063085 | 1.33 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr23_-_27589508 | 1.33 |
ENSDART00000178404
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr20_+_2460864 | 1.32 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr6_-_6254432 | 1.32 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr16_-_27224000 | 1.32 |
ENSDART00000126347
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr23_-_27101600 | 1.25 |
ENSDART00000139231
|
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr16_-_42303856 | 1.24 |
ENSDART00000180030
|
ppox
|
protoporphyrinogen oxidase |
| chr13_-_30149973 | 1.22 |
ENSDART00000041515
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr18_+_5543677 | 1.18 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr11_-_33960318 | 1.17 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr3_-_40658820 | 1.15 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr18_-_7448047 | 1.09 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr21_-_38581903 | 1.08 |
ENSDART00000148939
|
pof1b
|
POF1B, actin binding protein |
| chr12_+_20618522 | 1.07 |
ENSDART00000091452
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr5_-_72390259 | 1.07 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr20_+_33924235 | 1.07 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr17_+_28611746 | 1.00 |
ENSDART00000156711
ENSDART00000113300 |
mis18bp1
|
MIS18 binding protein 1 |
| chr3_+_23047241 | 0.99 |
ENSDART00000103858
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr11_+_7183025 | 0.98 |
ENSDART00000046670
ENSDART00000154009 ENSDART00000156974 ENSDART00000125619 |
thop1
|
thimet oligopeptidase 1 |
| chr15_-_37589600 | 0.95 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr13_-_30150592 | 0.95 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr9_+_2020667 | 0.93 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr13_+_35635672 | 0.89 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr11_+_1551603 | 0.89 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr21_-_11970199 | 0.89 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr23_-_31506854 | 0.88 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr12_+_19976400 | 0.86 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr3_-_10634438 | 0.85 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr20_+_51466046 | 0.85 |
ENSDART00000178065
|
tlr5b
|
toll-like receptor 5b |
| chr10_-_28193642 | 0.82 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr17_+_15213496 | 0.80 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr2_+_42072231 | 0.79 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr4_+_57099307 | 0.74 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr8_+_8973425 | 0.73 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr10_-_29101715 | 0.73 |
ENSDART00000149674
ENSDART00000171194 ENSDART00000192019 |
si:ch211-103f14.3
|
si:ch211-103f14.3 |
| chr5_-_38155005 | 0.73 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr15_+_24572926 | 0.73 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr3_-_34100700 | 0.72 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr6_-_9646275 | 0.71 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr24_-_7728409 | 0.69 |
ENSDART00000131392
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr9_-_52814204 | 0.68 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr14_-_46238186 | 0.68 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr16_+_1254390 | 0.67 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr15_-_1485086 | 0.64 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr5_+_26847190 | 0.62 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr2_+_45548890 | 0.62 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr12_+_46841918 | 0.59 |
ENSDART00000157464
|
adkb
|
adenosine kinase b |
| chr25_+_33063762 | 0.58 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr13_+_8987957 | 0.57 |
ENSDART00000148144
|
hcar1-3
|
hydroxycarboxylic acid receptor 1-3 |
| chr7_+_57109214 | 0.57 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr22_+_21972824 | 0.57 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr4_-_32458327 | 0.56 |
ENSDART00000182527
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr8_-_4431971 | 0.55 |
ENSDART00000141728
ENSDART00000133519 |
rad9b
|
RAD9 checkpoint clamp component B |
| chr15_-_47812268 | 0.53 |
ENSDART00000190231
|
tomt
|
transmembrane O-methyltransferase |
| chr7_+_38510197 | 0.53 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr7_+_29512673 | 0.53 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr9_+_21281652 | 0.53 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr25_+_9027831 | 0.52 |
ENSDART00000155855
|
im:7145024
|
im:7145024 |
| chr15_-_1484795 | 0.52 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr11_+_42765963 | 0.51 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr9_-_18716 | 0.50 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr2_-_3077027 | 0.49 |
ENSDART00000109319
|
arf1
|
ADP-ribosylation factor 1 |
| chr10_-_244745 | 0.49 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr9_+_29985010 | 0.48 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr15_-_34668485 | 0.48 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr17_+_23255365 | 0.47 |
ENSDART00000180277
|
AL935174.5
|
|
| chr5_-_1218801 | 0.46 |
ENSDART00000184398
|
CABZ01048143.1
|
|
| chr2_-_37684641 | 0.45 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr21_+_3928947 | 0.45 |
ENSDART00000149777
|
setx
|
senataxin |
| chr10_-_35410518 | 0.45 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr22_+_19311411 | 0.44 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr4_+_20177526 | 0.43 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr21_-_38582061 | 0.43 |
ENSDART00000193891
ENSDART00000124349 |
pof1b
|
POF1B, actin binding protein |
| chr5_+_61476014 | 0.43 |
ENSDART00000050906
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr15_+_23356600 | 0.43 |
ENSDART00000113279
|
rnf26
|
ring finger protein 26 |
| chr8_-_49201258 | 0.43 |
ENSDART00000114584
ENSDART00000142172 |
zgc:171501
|
zgc:171501 |
| chr14_-_21238046 | 0.42 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr2_-_14793343 | 0.42 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr2_+_2967255 | 0.41 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr12_+_20618336 | 0.41 |
ENSDART00000084007
|
si:dkey-220f10.4
|
si:dkey-220f10.4 |
| chr4_-_32458542 | 0.41 |
ENSDART00000127211
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_14743655 | 0.41 |
ENSDART00000192518
|
BX571719.1
|
|
| chr14_-_8940499 | 0.40 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr14_+_20351 | 0.39 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr19_+_30884706 | 0.39 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr4_-_68868229 | 0.38 |
ENSDART00000159219
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr20_-_49657134 | 0.38 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr10_+_40774215 | 0.37 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr21_+_3021702 | 0.37 |
ENSDART00000188235
|
CABZ01023247.1
|
|
| chr5_-_29112956 | 0.35 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr22_-_19102256 | 0.34 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr12_+_48683208 | 0.32 |
ENSDART00000188582
|
mmp21
|
matrix metallopeptidase 21 |
| chr23_+_45734011 | 0.31 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr21_+_43172506 | 0.31 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr20_+_54037138 | 0.30 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr16_+_41570653 | 0.30 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr21_+_41839443 | 0.29 |
ENSDART00000010942
|
rnf14
|
ring finger protein 14 |
| chr17_+_51224421 | 0.29 |
ENSDART00000025229
|
adi1
|
acireductone dioxygenase 1 |
| chr4_-_76205022 | 0.28 |
ENSDART00000159687
ENSDART00000187593 |
si:ch211-106j21.5
|
si:ch211-106j21.5 |
| chr24_+_30392834 | 0.28 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr11_-_18017918 | 0.28 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr25_+_35133404 | 0.27 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr4_+_49196098 | 0.26 |
ENSDART00000150678
|
si:dkey-40n15.1
|
si:dkey-40n15.1 |
| chr17_-_51224800 | 0.25 |
ENSDART00000150089
|
psen1
|
presenilin 1 |
| chr21_+_21741804 | 0.24 |
ENSDART00000112068
|
defbl1
|
defensin, beta-like 1 |
| chr2_-_36602952 | 0.23 |
ENSDART00000165032
|
tradv40.0
|
T-cell receptor alpha/delta variable 40.0 |
| chr9_-_11549379 | 0.23 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr10_-_18463934 | 0.21 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr5_+_33488860 | 0.20 |
ENSDART00000135571
|
si:dkey-238j22.1
|
si:dkey-238j22.1 |
| chr5_+_64277604 | 0.19 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr4_+_61840440 | 0.19 |
ENSDART00000169427
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_74892355 | 0.18 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr9_+_14023386 | 0.18 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr2_-_50126337 | 0.17 |
ENSDART00000009347
|
exoc3
|
exocyst complex component 3 |
| chr23_-_24542156 | 0.17 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr16_-_14552199 | 0.16 |
ENSDART00000133368
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr3_+_22375596 | 0.14 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr22_-_10586191 | 0.14 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr13_+_22421151 | 0.14 |
ENSDART00000122687
|
opn4a
|
opsin 4a (melanopsin) |
| chr1_+_58066109 | 0.14 |
ENSDART00000115190
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr4_-_71151245 | 0.13 |
ENSDART00000165415
|
si:dkey-193i10.4
|
si:dkey-193i10.4 |
| chr2_+_42072689 | 0.12 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr19_-_19379084 | 0.12 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr4_+_53119675 | 0.12 |
ENSDART00000167470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr4_+_54618332 | 0.10 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr18_+_7543347 | 0.10 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr13_+_30215765 | 0.09 |
ENSDART00000041026
ENSDART00000133049 |
qrfpra
|
pyroglutamylated RFamide peptide receptor a |
| chr5_+_23994038 | 0.09 |
ENSDART00000186175
|
AL928875.1
|
|
| chr12_-_46315898 | 0.09 |
ENSDART00000153138
|
si:ch211-226h7.3
|
si:ch211-226h7.3 |
| chr14_+_39156 | 0.07 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr7_+_13491452 | 0.06 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr13_+_18523661 | 0.05 |
ENSDART00000034852
|
tlr4bb
|
toll-like receptor 4b, duplicate b |
| chr13_-_31166544 | 0.05 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr25_-_2355107 | 0.04 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr2_+_42871831 | 0.04 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr7_+_22792132 | 0.04 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr9_-_14992730 | 0.04 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr4_-_77377596 | 0.03 |
ENSDART00000186068
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr10_+_2876020 | 0.03 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr19_-_32042105 | 0.03 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 1.3 | 7.9 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.5 | 2.2 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.5 | 2.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 7.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 6.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 0.9 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.3 | 1.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.3 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 1.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 1.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.9 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.8 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 1.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 1.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 1.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.4 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.9 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 3.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 1.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.3 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.0 | 0.5 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.5 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 1.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 1.2 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 2.5 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 2.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 0.9 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.3 | 1.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.0 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 8.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.5 | 5.9 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 1.1 | 3.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.7 | 7.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.4 | 1.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 1.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 1.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 2.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.9 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 0.5 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.8 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.7 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |