Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
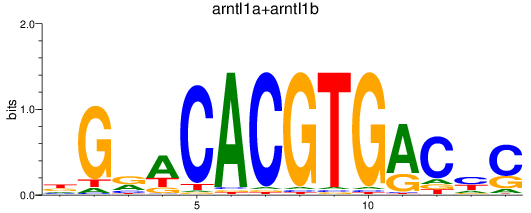
Results for arntl1a+arntl1b
Z-value: 0.91
Transcription factors associated with arntl1a+arntl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arntl1a
|
ENSDARG00000006791 | aryl hydrocarbon receptor nuclear translocator-like 1a |
|
arntl1b
|
ENSDARG00000035732 | aryl hydrocarbon receptor nuclear translocator-like 1b |
|
arntl1b
|
ENSDARG00000114562 | aryl hydrocarbon receptor nuclear translocator-like 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arntl1b | dr11_v1_chr7_+_66072580_66072580 | 0.17 | 1.0e-01 | Click! |
| arntl1a | dr11_v1_chr25_-_17910714_17910714 | 0.02 | 8.7e-01 | Click! |
Activity profile of arntl1a+arntl1b motif
Sorted Z-values of arntl1a+arntl1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arntl1a+arntl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.8 | 3.8 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.7 | 7.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.5 | 5.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.5 | 1.6 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.5 | 1.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.4 | 1.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.4 | 6.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 5.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 5.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 3.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 0.6 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 1.1 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.5 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 0.7 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.6 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.7 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 6.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.0 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 1.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.6 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.6 | GO:0030194 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 5.1 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 2.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.0 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 1.2 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.5 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.1 | GO:0030388 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 1.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 16.1 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 1.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.8 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.5 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 3.0 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 2.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.7 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 1.6 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.5 | 2.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.4 | 5.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 1.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 1.3 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0030132 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 8.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.6 | 1.7 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.4 | 1.3 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.4 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 1.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 4.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 5.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 17.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 16.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 6.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 3.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 5.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 4.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |