Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
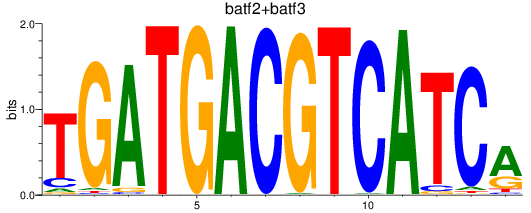
Results for batf2+batf3
Z-value: 0.83
Transcription factors associated with batf2+batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
batf3
|
ENSDARG00000042577 | basic leucine zipper transcription factor, ATF-like 3 |
|
batf2
|
ENSDARG00000105562 | si_dkey-23i12.7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-23i12.7 | dr11_v1_chr7_+_25053331_25053331 | 0.23 | 2.4e-02 | Click! |
| batf3 | dr11_v1_chr20_-_37813863_37813863 | 0.13 | 2.1e-01 | Click! |
Activity profile of batf2+batf3 motif
Sorted Z-values of batf2+batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_20637866 | 10.51 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr16_-_12173554 | 10.02 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr20_+_20638034 | 9.56 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr7_+_40228422 | 9.44 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr22_+_1796057 | 8.45 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr14_+_11950011 | 6.52 |
ENSDART00000188138
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr11_-_29623380 | 6.34 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr13_+_36146415 | 6.24 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr21_-_23308286 | 6.11 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr1_+_10305611 | 5.76 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr15_-_762319 | 5.41 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr17_+_29345606 | 5.10 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr22_+_1904546 | 4.41 |
ENSDART00000164865
|
si:dkey-15h8.10
|
si:dkey-15h8.10 |
| chr22_+_2801464 | 4.15 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr22_+_2715815 | 4.10 |
ENSDART00000042770
|
znf1163
|
zinc finger protein 1163 |
| chr22_+_2120815 | 4.07 |
ENSDART00000169133
ENSDART00000166644 ENSDART00000162221 ENSDART00000157840 |
znf1155
|
zinc finger protein 1155 |
| chr22_+_2819613 | 3.86 |
ENSDART00000131234
|
si:dkey-20i20.3
|
si:dkey-20i20.3 |
| chr22_+_1922293 | 3.84 |
ENSDART00000165758
|
znf1181
|
zinc finger protein 1181 |
| chr22_+_1940595 | 3.84 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr24_+_744713 | 3.82 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr22_+_1700552 | 3.72 |
ENSDART00000166185
|
znf1154
|
zinc finger protein 1154 |
| chr22_+_2183110 | 3.70 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr22_+_2345752 | 3.60 |
ENSDART00000143687
|
zgc:174224
|
zgc:174224 |
| chr22_+_2844865 | 3.53 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr22_+_2254972 | 3.46 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr11_-_44163164 | 3.42 |
ENSDART00000047126
|
clcn4
|
chloride channel, voltage-sensitive 4 |
| chr13_-_40754499 | 3.35 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr16_+_27614989 | 3.35 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr14_+_33264303 | 3.23 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr22_+_2778600 | 3.21 |
ENSDART00000134028
|
si:dkey-20i20.9
|
si:dkey-20i20.9 |
| chr18_-_46063773 | 3.21 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr5_+_11812089 | 3.18 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr22_+_2524615 | 3.13 |
ENSDART00000134277
|
znf1003
|
zinc finger protein 1003 |
| chr22_+_1930589 | 3.05 |
ENSDART00000159807
|
znf1153
|
zinc finger protein 1153 |
| chr22_+_1853999 | 2.87 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr22_+_2111120 | 2.75 |
ENSDART00000165525
|
znf1144
|
zinc finger protein 1144 |
| chr1_+_41666611 | 2.72 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr22_+_1641547 | 2.71 |
ENSDART00000159804
|
si:dkey-1b17.2
|
si:dkey-1b17.2 |
| chr17_+_32500387 | 2.68 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr22_+_2648931 | 2.67 |
ENSDART00000106357
|
zgc:110821
|
zgc:110821 |
| chr22_+_1947494 | 2.60 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr22_+_2298419 | 2.58 |
ENSDART00000143499
|
znf1180
|
zinc finger protein 1180 |
| chr22_+_2207502 | 2.52 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr22_+_1911269 | 2.51 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr22_+_30330574 | 2.50 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr8_+_53120278 | 2.46 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr22_+_1517318 | 2.36 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr22_+_1953096 | 2.34 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr22_+_2239254 | 2.32 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr15_-_20412286 | 2.30 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr17_+_15674052 | 2.30 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_+_2247645 | 2.09 |
ENSDART00000143366
ENSDART00000147852 |
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr25_+_7321675 | 2.03 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr22_+_2228919 | 1.92 |
ENSDART00000133475
|
znf1161
|
zinc finger protein 1161 |
| chr22_+_1462177 | 1.85 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr22_+_2189356 | 1.83 |
ENSDART00000157502
ENSDART00000164188 |
znf1165
|
zinc finger protein 1165 |
| chr14_-_32959851 | 1.76 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr22_+_2390544 | 1.70 |
ENSDART00000147345
ENSDART00000133611 |
zgc:171435
|
zgc:171435 |
| chr4_+_30775376 | 1.62 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr21_-_14762944 | 1.62 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr20_-_21994901 | 1.60 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr16_+_5184402 | 1.53 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr22_+_2315996 | 1.49 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr2_+_27341559 | 1.40 |
ENSDART00000176838
|
tesk2
|
testis-specific kinase 2 |
| chr16_-_54942532 | 1.33 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr7_+_29301497 | 1.33 |
ENSDART00000099327
|
rab8b
|
RAB8B, member RAS oncogene family |
| chr22_+_1821718 | 1.26 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr22_+_2751887 | 1.23 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr11_+_28166165 | 1.16 |
ENSDART00000169360
ENSDART00000192311 |
ephb2b
|
eph receptor B2b |
| chr22_+_2403068 | 1.03 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr22_+_1668471 | 0.83 |
ENSDART00000163358
|
si:dkey-1b17.5
|
si:dkey-1b17.5 |
| chr5_+_37035978 | 0.77 |
ENSDART00000167418
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr8_-_21110233 | 0.54 |
ENSDART00000127371
ENSDART00000100276 |
tmco1
|
transmembrane and coiled-coil domains 1 |
| chr4_+_70556298 | 0.38 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr7_-_69352424 | 0.33 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr1_-_20068155 | 0.26 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr21_-_23307653 | 0.24 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr24_+_25822999 | 0.18 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_-_16413606 | 0.16 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr16_-_41667101 | 0.15 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr5_+_20823409 | 0.06 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr22_+_1751640 | 0.04 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
Network of associatons between targets according to the STRING database.
First level regulatory network of batf2+batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.6 | 3.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 6.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 5.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.5 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 6.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.3 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.5 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 1.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 9.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 85.5 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
| 0.0 | 3.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 12.7 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.0 | 1.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.6 | GO:0060026 | convergent extension(GO:0060026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 3.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 8.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 14.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.3 | 1.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 3.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 5.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 6.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 6.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 10.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 9.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 90.4 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.7 | GO:0004497 | monooxygenase activity(GO:0004497) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |