Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
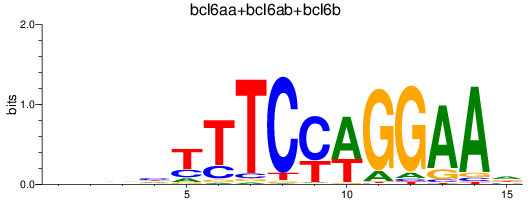
Results for bcl6aa+bcl6ab+bcl6b
Z-value: 0.86
Transcription factors associated with bcl6aa+bcl6ab+bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl6ab
|
ENSDARG00000069295 | BCL6A transcription repressor b |
|
bcl6b
|
ENSDARG00000069335 | BCL6B transcription repressor |
|
bcl6aa
|
ENSDARG00000070864 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000111395 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000112502 | BCL6A transcription repressor a |
|
bcl6aa
|
ENSDARG00000116260 | BCL6A transcription repressor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl6ab | dr11_v1_chr2_-_10098191_10098191 | 0.17 | 1.1e-01 | Click! |
| bcl6a | dr11_v1_chr6_-_28222592_28222592 | -0.12 | 2.5e-01 | Click! |
| bcl6b | dr11_v1_chr7_+_20017211_20017211 | -0.08 | 4.6e-01 | Click! |
Activity profile of bcl6aa+bcl6ab+bcl6b motif
Sorted Z-values of bcl6aa+bcl6ab+bcl6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_11457500 | 10.68 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr16_-_36834505 | 10.08 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr11_-_8167799 | 9.01 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr5_-_63509581 | 8.08 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr25_-_17395315 | 7.07 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr15_+_20239141 | 6.78 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr5_+_28849155 | 6.76 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr12_-_4243268 | 6.55 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr21_-_26114886 | 6.52 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr20_+_23440632 | 6.35 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr20_+_25581627 | 6.32 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr16_-_45917322 | 6.20 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr3_-_57737913 | 5.89 |
ENSDART00000113309
|
LGALS3BP (1 of many)
|
zgc:112492 |
| chr19_-_5351980 | 5.60 |
ENSDART00000163304
ENSDART00000027701 |
krt92
|
keratin 92 |
| chr24_-_2829049 | 5.56 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr3_+_19299309 | 5.46 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr5_+_28858345 | 4.89 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr16_-_27749172 | 4.41 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr3_+_49021079 | 4.30 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr15_-_23793641 | 4.29 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr20_+_25552057 | 4.23 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr20_-_25551676 | 4.10 |
ENSDART00000063081
|
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr23_+_42292748 | 4.10 |
ENSDART00000166113
ENSDART00000158684 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr11_+_14286160 | 4.05 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr5_+_24179307 | 3.95 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr4_-_12323228 | 3.93 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr6_-_8498908 | 3.89 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_+_43375326 | 3.87 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr8_+_1189798 | 3.76 |
ENSDART00000193474
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr18_+_18863167 | 3.63 |
ENSDART00000091094
|
pllp
|
plasmolipin |
| chr18_-_14860435 | 3.63 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr8_+_47099033 | 3.58 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr23_+_42346799 | 3.25 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr8_+_22359881 | 3.24 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr7_-_19168375 | 3.22 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr18_+_18982077 | 3.19 |
ENSDART00000006300
|
hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr13_+_33688474 | 3.15 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr6_-_8498676 | 3.11 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr17_+_10318071 | 3.00 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr20_-_33704753 | 2.94 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr2_-_10703621 | 2.84 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr25_-_29415369 | 2.82 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr3_-_44059902 | 2.69 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr24_-_33291784 | 2.62 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr3_-_50998577 | 2.61 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr14_-_11507211 | 2.53 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr7_+_61764040 | 2.49 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr18_+_44532370 | 2.48 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr7_+_14005111 | 2.37 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr3_+_3139240 | 2.37 |
ENSDART00000105014
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr2_-_11120220 | 2.36 |
ENSDART00000184793
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr7_+_37372479 | 2.33 |
ENSDART00000173652
|
sall1a
|
spalt-like transcription factor 1a |
| chr21_-_17956416 | 2.32 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr7_+_26549846 | 2.32 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr22_-_15578402 | 2.28 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr22_+_7480465 | 2.26 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr1_-_46663997 | 2.23 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr21_+_12010505 | 2.22 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr22_-_16180467 | 2.17 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr4_-_18436899 | 2.13 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr2_+_35595454 | 2.09 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr9_+_40825065 | 2.08 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr16_+_42772678 | 2.07 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr5_-_29195063 | 2.06 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr7_+_26167420 | 2.06 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr11_-_287670 | 2.05 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr17_+_34206167 | 2.04 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr18_+_22109379 | 2.00 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr15_-_20709289 | 1.99 |
ENSDART00000136767
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr20_-_19858936 | 1.97 |
ENSDART00000161579
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr8_-_23240156 | 1.95 |
ENSDART00000131632
|
ptk6a
|
PTK6 protein tyrosine kinase 6a |
| chr6_+_28294113 | 1.94 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_-_23365737 | 1.94 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr22_+_26400519 | 1.90 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr3_-_44012748 | 1.88 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr19_-_35428815 | 1.87 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr20_-_40367493 | 1.84 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr15_-_3282220 | 1.81 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr7_-_54320088 | 1.80 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr8_+_23711842 | 1.79 |
ENSDART00000128783
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr8_-_21091961 | 1.75 |
ENSDART00000100281
|
aldh9a1a.2
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 2 |
| chr10_-_3332362 | 1.75 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr2_-_37465517 | 1.74 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr9_-_17417628 | 1.74 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr24_+_11381400 | 1.74 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr18_-_46183462 | 1.73 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr25_+_11281970 | 1.71 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr17_-_23631400 | 1.69 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr20_+_52458765 | 1.69 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr20_-_23656516 | 1.66 |
ENSDART00000149735
|
cbr4
|
carbonyl reductase 4 |
| chr19_+_4038589 | 1.64 |
ENSDART00000169271
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr1_-_40208237 | 1.64 |
ENSDART00000191987
|
AL953865.1
|
|
| chr18_+_44532199 | 1.61 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr5_+_50953240 | 1.61 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr16_-_26296477 | 1.60 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr6_-_40352215 | 1.60 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr4_+_47257854 | 1.59 |
ENSDART00000173868
|
crestin
|
crestin |
| chr20_+_25625872 | 1.53 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr21_-_30026359 | 1.53 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr19_-_3821678 | 1.53 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr23_+_45229198 | 1.51 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr17_-_30521043 | 1.50 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr9_+_20869166 | 1.49 |
ENSDART00000147892
|
wdr3
|
WD repeat domain 3 |
| chr19_+_32856907 | 1.48 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr7_+_26029672 | 1.48 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr17_+_16046314 | 1.45 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_-_26026956 | 1.45 |
ENSDART00000102346
|
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr8_-_41228530 | 1.44 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr13_-_37122217 | 1.43 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr6_-_35106425 | 1.42 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr18_+_45666489 | 1.41 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr19_-_8798178 | 1.39 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr25_-_34280080 | 1.38 |
ENSDART00000085251
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_+_10318089 | 1.37 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr1_-_9195629 | 1.36 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr2_+_111919 | 1.36 |
ENSDART00000149391
|
fggy
|
FGGY carbohydrate kinase domain containing |
| chr17_+_16046132 | 1.35 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_+_73215536 | 1.35 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr3_+_32492467 | 1.33 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr3_+_60589157 | 1.33 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr9_-_9225980 | 1.32 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr25_-_21822426 | 1.32 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr16_-_42965192 | 1.31 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr14_+_26229056 | 1.31 |
ENSDART00000179045
|
LO018208.1
|
|
| chr16_-_41667101 | 1.30 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr1_-_44928987 | 1.29 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr2_-_59327299 | 1.29 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr13_-_11967769 | 1.28 |
ENSDART00000158369
|
ARL3 (1 of many)
|
zgc:110197 |
| chr5_+_9360394 | 1.28 |
ENSDART00000124642
|
FP236810.2
|
|
| chr3_+_1698430 | 1.27 |
ENSDART00000193625
|
BX321875.4
|
|
| chr15_+_19324697 | 1.23 |
ENSDART00000022015
|
vps26b
|
VPS26 retromer complex component B |
| chr7_-_34434889 | 1.21 |
ENSDART00000159982
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr12_+_5358409 | 1.20 |
ENSDART00000152632
|
plce1
|
phospholipase C, epsilon 1 |
| chr17_-_12498096 | 1.20 |
ENSDART00000149551
ENSDART00000105215 ENSDART00000191207 |
emilin1b
|
elastin microfibril interfacer 1b |
| chr7_+_32369026 | 1.20 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr18_-_44888375 | 1.19 |
ENSDART00000160506
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr13_-_47554194 | 1.18 |
ENSDART00000181508
|
acoxl
|
acyl-CoA oxidase-like |
| chr4_+_57580303 | 1.16 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr9_-_43644261 | 1.16 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr24_+_7828097 | 1.15 |
ENSDART00000134975
|
zgc:101569
|
zgc:101569 |
| chr21_+_26073104 | 1.15 |
ENSDART00000193273
|
rpl23a
|
ribosomal protein L23a |
| chr18_+_13315739 | 1.12 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr5_+_1911814 | 1.10 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr16_+_46294337 | 1.10 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr8_-_23783633 | 1.09 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr25_+_16915974 | 1.07 |
ENSDART00000188923
|
zgc:77158
|
zgc:77158 |
| chr5_+_19494198 | 1.07 |
ENSDART00000006673
|
mvk
|
mevalonate kinase |
| chr1_-_40189893 | 1.06 |
ENSDART00000133738
|
si:ch211-113e8.6
|
si:ch211-113e8.6 |
| chr4_+_49322310 | 1.06 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr11_+_36409457 | 1.05 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr6_+_47843760 | 1.01 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr22_-_37611681 | 1.00 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr17_-_48743076 | 0.98 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr5_-_51166025 | 0.95 |
ENSDART00000097471
|
card9
|
caspase recruitment domain family, member 9 |
| chr2_+_3428357 | 0.95 |
ENSDART00000125967
|
CU633991.1
|
|
| chr18_+_14684115 | 0.94 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr12_-_26558038 | 0.94 |
ENSDART00000039510
|
tap2t
|
transporter associated with antigen processing, subunit type t, teleost specific |
| chr1_-_56596701 | 0.94 |
ENSDART00000133693
|
LO018605.1
|
|
| chr20_+_21595244 | 0.93 |
ENSDART00000010643
|
esr2a
|
estrogen receptor 2a |
| chr9_-_38579758 | 0.92 |
ENSDART00000131738
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr21_+_45626136 | 0.91 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr23_-_19715799 | 0.90 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr16_-_4610255 | 0.90 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr17_+_24597001 | 0.90 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr20_+_4793790 | 0.89 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr21_-_27338639 | 0.88 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr22_-_27706576 | 0.88 |
ENSDART00000188564
|
CR547131.1
|
|
| chr12_-_44151296 | 0.87 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr1_-_57839070 | 0.87 |
ENSDART00000152571
|
si:dkey-1c7.3
|
si:dkey-1c7.3 |
| chr3_-_54524194 | 0.86 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr13_-_37631092 | 0.86 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr23_-_14830627 | 0.86 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr21_+_27297145 | 0.84 |
ENSDART00000146588
|
si:dkey-175m17.6
|
si:dkey-175m17.6 |
| chr7_-_24373662 | 0.84 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr3_-_55139127 | 0.83 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr4_+_57099307 | 0.83 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr9_+_26103814 | 0.83 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr1_+_58139102 | 0.82 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr17_-_50430817 | 0.82 |
ENSDART00000154007
|
si:ch211-235i11.4
|
si:ch211-235i11.4 |
| chr20_+_27087539 | 0.82 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr13_-_50234122 | 0.81 |
ENSDART00000164338
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr5_-_35200590 | 0.80 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr5_-_41841675 | 0.80 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr3_-_30888415 | 0.79 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr4_+_6639292 | 0.79 |
ENSDART00000158573
|
SMIM30
|
si:dkey-112e7.2 |
| chr22_-_3344613 | 0.78 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr16_+_40954481 | 0.77 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr3_-_3703572 | 0.77 |
ENSDART00000111017
|
si:ch211-163m16.7
|
si:ch211-163m16.7 |
| chr6_-_14004772 | 0.77 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr6_+_41503854 | 0.76 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr14_-_5678457 | 0.76 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr19_-_6631900 | 0.76 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr18_-_21170264 | 0.74 |
ENSDART00000175265
|
BX571884.1
|
|
| chr7_-_12869545 | 0.74 |
ENSDART00000163045
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr1_+_55600504 | 0.74 |
ENSDART00000123946
|
umod
|
uromodulin |
| chr10_-_39281475 | 0.73 |
ENSDART00000175136
|
cry5
|
cryptochrome circadian clock 5 |
| chr4_-_13518381 | 0.73 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr20_+_27646772 | 0.72 |
ENSDART00000141697
|
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr7_+_73295890 | 0.71 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr7_+_32369463 | 0.71 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl6aa+bcl6ab+bcl6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 2.3 | 7.0 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 1.1 | 4.6 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 1.1 | 4.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.9 | 3.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.9 | 5.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.7 | 2.1 | GO:0002857 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.6 | 1.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.6 | 4.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 28.5 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.5 | 2.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.5 | 2.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.5 | 1.8 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 1.3 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.4 | 1.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.4 | 1.7 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.4 | 1.2 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.4 | 6.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 1.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.4 | 2.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 2.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 3.2 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.3 | 1.9 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.3 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 3.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.3 | 8.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.3 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.3 | 1.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 1.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 0.8 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 2.6 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 1.3 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.0 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 2.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 1.0 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.2 | 1.8 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 2.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 3.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 4.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.7 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.6 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.2 | 2.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 2.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 2.6 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 3.0 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 11.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 3.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 1.5 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.7 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.5 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.9 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.6 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 3.7 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 0.9 | GO:0002831 | regulation of response to biotic stimulus(GO:0002831) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.8 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 1.0 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.2 | GO:1905067 | negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.8 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 1.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 2.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 9.0 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 1.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0035474 | selective angioblast sprouting(GO:0035474) angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.6 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.4 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 7.9 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.0 | 3.2 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.5 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.7 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 1.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 6.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 2.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.2 | GO:0045334 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 4.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 5.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 5.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 12.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 5.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.7 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 15.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0044440 | endosomal part(GO:0044440) |
| 0.0 | 1.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.4 | 5.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.1 | 9.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.1 | 10.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.1 | 4.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.9 | 3.8 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.8 | 3.2 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.8 | 2.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.7 | 3.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.7 | 2.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.6 | 2.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 1.6 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.5 | 2.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 28.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.4 | 1.7 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.4 | 1.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 5.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 2.9 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 1.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.3 | 6.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.0 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.3 | 2.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 0.9 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.3 | 2.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.3 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 0.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 2.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.7 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.2 | 3.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 2.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 2.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 1.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.7 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.9 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.8 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.6 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 20.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 9.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 6.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.5 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 12.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 3.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.3 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 2.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.0 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 2.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 0.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 3.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 2.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 8.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 3.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 3.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 5.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |