Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
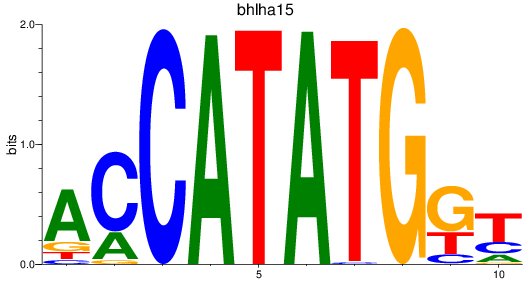
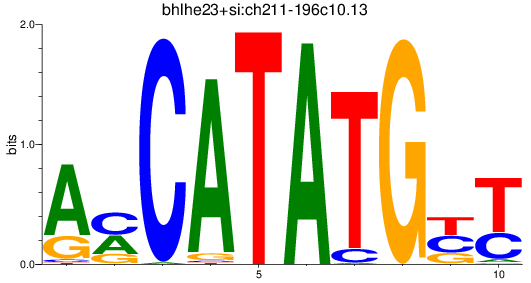
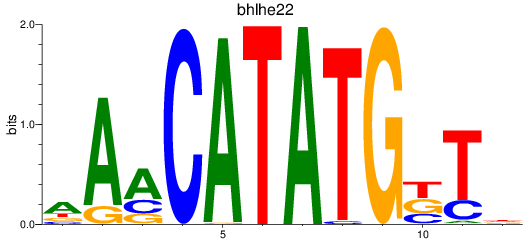
Results for bhlha15_bhlhe23+si:ch211-196c10.13_bhlhe22
Z-value: 0.92
Transcription factors associated with bhlha15_bhlhe23+si:ch211-196c10.13_bhlhe22
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bhlha15
|
ENSDARG00000045166 | basic helix-loop-helix family, member a15 |
|
bhlhe23
|
ENSDARG00000037588 | basic helix-loop-helix family, member e23 |
|
si_ch211-196c10.13
|
ENSDARG00000096756 | si_ch211-196c10.13 |
|
bhlhe22
|
ENSDARG00000058039 | basic helix-loop-helix family, member e22 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bhlhe23 | dr11_v1_chr23_-_17657348_17657348 | 0.79 | 1.3e-21 | Click! |
| bhlhe22 | dr11_v1_chr24_+_24461558_24461558 | 0.79 | 6.0e-21 | Click! |
| si:ch211-196c10.13 | dr11_v1_chr8_+_23142946_23142946 | 0.67 | 2.3e-13 | Click! |
| bhlha15 | dr11_v1_chr12_+_17754859_17754859 | 0.01 | 9.2e-01 | Click! |
Activity profile of bhlha15_bhlhe23+si:ch211-196c10.13_bhlhe22 motif
Sorted Z-values of bhlha15_bhlhe23+si:ch211-196c10.13_bhlhe22 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_22012389 | 24.92 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr4_-_8882572 | 18.66 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr12_-_14922955 | 15.36 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr2_-_27775236 | 15.35 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr14_+_25817246 | 14.77 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr1_-_16507812 | 14.04 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr21_+_11468642 | 13.73 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr5_-_27994679 | 13.31 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr14_-_4682114 | 12.78 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr6_+_54142311 | 10.96 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr19_-_7406933 | 10.84 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr23_-_21453614 | 10.39 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr4_+_15968483 | 9.34 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr13_-_24311628 | 9.20 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr13_+_31285660 | 9.16 |
ENSDART00000148352
|
gdf10a
|
growth differentiation factor 10a |
| chr19_-_21766461 | 8.35 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr23_+_21544227 | 8.18 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr10_-_31440500 | 7.81 |
ENSDART00000024778
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr14_+_31618982 | 7.79 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr20_-_31252809 | 7.32 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr14_-_4556896 | 6.67 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr13_+_11439486 | 6.52 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr24_+_2801765 | 6.50 |
ENSDART00000172385
|
LYRM4
|
si:ch211-152c8.4 |
| chr14_+_25816874 | 6.41 |
ENSDART00000005499
|
glra1
|
glycine receptor, alpha 1 |
| chr17_-_42218652 | 6.13 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr23_+_23658474 | 5.94 |
ENSDART00000162838
|
agrn
|
agrin |
| chr20_+_34326874 | 5.85 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr21_-_25601648 | 5.76 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr12_+_24562667 | 5.27 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr13_+_28821841 | 5.04 |
ENSDART00000179900
|
CU639469.1
|
|
| chr2_-_5074812 | 4.96 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr13_+_15581270 | 4.81 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr7_-_31938938 | 4.79 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr19_-_15855427 | 4.73 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr15_+_15856178 | 4.65 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr8_-_39952727 | 4.60 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr3_+_50310684 | 4.09 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr13_-_31441042 | 3.61 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr22_+_11857356 | 3.55 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr3_+_28953274 | 3.37 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr4_+_20318127 | 3.09 |
ENSDART00000028856
ENSDART00000132909 |
cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr14_+_23874062 | 3.02 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr12_-_3753131 | 2.96 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr2_-_42558549 | 2.69 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr1_-_16394814 | 2.57 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr19_+_1184878 | 2.48 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr7_-_15413019 | 2.34 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr5_+_45138934 | 2.34 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr24_-_9689915 | 2.32 |
ENSDART00000185972
ENSDART00000093046 |
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr11_-_24532988 | 2.27 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr1_+_42874410 | 2.13 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr22_+_3238474 | 2.12 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr13_+_31286076 | 2.00 |
ENSDART00000142725
|
gdf10a
|
growth differentiation factor 10a |
| chr19_+_31904836 | 1.94 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr4_+_12617108 | 1.93 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr17_+_28882977 | 1.86 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr14_+_36521553 | 1.81 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr4_+_14360372 | 1.80 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr11_-_34521342 | 1.79 |
ENSDART00000114004
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr24_+_26337623 | 1.74 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr3_+_47322494 | 1.73 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr14_-_31854830 | 1.68 |
ENSDART00000148550
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr23_-_4019699 | 1.61 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr14_+_40852497 | 1.61 |
ENSDART00000128588
ENSDART00000166065 |
taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_-_22568203 | 1.52 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr11_+_16152316 | 1.48 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr5_+_36693859 | 1.47 |
ENSDART00000019259
|
dlb
|
deltaB |
| chr7_-_50395395 | 1.47 |
ENSDART00000065868
|
vps33b
|
vacuolar protein sorting 33B |
| chr12_-_35095414 | 1.37 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr9_+_38168012 | 1.37 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr15_-_14884332 | 1.36 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr7_-_50395059 | 1.34 |
ENSDART00000191150
|
vps33b
|
vacuolar protein sorting 33B |
| chr14_-_12071679 | 1.31 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr18_+_12147971 | 1.30 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr14_-_12071447 | 1.29 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr11_+_5499661 | 1.27 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr15_+_4988189 | 1.25 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr4_+_5796761 | 1.24 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr10_+_43088270 | 1.21 |
ENSDART00000191221
|
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr1_+_5275811 | 1.20 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr4_-_27129697 | 1.20 |
ENSDART00000131240
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr6_+_7553085 | 1.16 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr24_+_17140938 | 1.16 |
ENSDART00000149134
|
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr10_-_23358357 | 1.13 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr19_-_24125457 | 1.10 |
ENSDART00000080632
|
zgc:64022
|
zgc:64022 |
| chr12_+_17603528 | 1.10 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr23_+_31979602 | 1.04 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_-_52674089 | 1.02 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr1_-_17711636 | 0.94 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr3_+_35406998 | 0.91 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr2_+_33383227 | 0.90 |
ENSDART00000183068
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr14_-_30897177 | 0.89 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr2_-_32551178 | 0.89 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr14_+_21159693 | 0.85 |
ENSDART00000192678
|
zgc:136929
|
zgc:136929 |
| chr21_-_13751535 | 0.84 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr19_-_17864213 | 0.84 |
ENSDART00000151043
|
ints8
|
integrator complex subunit 8 |
| chr22_-_18116635 | 0.83 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr17_+_12658411 | 0.79 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr9_-_6502491 | 0.77 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr10_-_7347311 | 0.77 |
ENSDART00000168885
|
nrg1
|
neuregulin 1 |
| chr5_+_37379825 | 0.74 |
ENSDART00000171826
|
klhl13
|
kelch-like family member 13 |
| chr6_-_36182115 | 0.71 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr22_-_14475927 | 0.70 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr3_+_43774369 | 0.70 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr20_+_3997684 | 0.69 |
ENSDART00000113184
|
arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr18_+_39327010 | 0.68 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr3_+_35406395 | 0.65 |
ENSDART00000055266
ENSDART00000150918 |
rbbp6
|
retinoblastoma binding protein 6 |
| chr8_-_51507144 | 0.62 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr7_+_41421998 | 0.60 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr13_+_30421472 | 0.60 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr11_-_16152105 | 0.58 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr15_+_32297441 | 0.55 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr8_-_22660678 | 0.52 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr22_-_24248420 | 0.52 |
ENSDART00000165433
|
rgs2
|
regulator of G protein signaling 2 |
| chr16_-_39267185 | 0.47 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr9_-_34121839 | 0.46 |
ENSDART00000000004
|
slc35a5
|
solute carrier family 35, member A5 |
| chr13_+_22295905 | 0.44 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr11_+_6684968 | 0.42 |
ENSDART00000139454
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr17_-_51260476 | 0.41 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr19_+_38167468 | 0.37 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr4_+_9178913 | 0.36 |
ENSDART00000168558
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr16_-_45225520 | 0.35 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr24_+_17345521 | 0.33 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr2_-_39036604 | 0.30 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr9_+_32860345 | 0.30 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr20_+_19423823 | 0.29 |
ENSDART00000152216
|
znf513a
|
zinc finger protein 513a |
| chr1_-_17711361 | 0.29 |
ENSDART00000078848
|
ufsp2
|
ufm1-specific peptidase 2 |
| chr20_-_1265562 | 0.27 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr5_-_41709234 | 0.20 |
ENSDART00000083656
|
atxn2
|
ataxin 2 |
| chr19_-_7540821 | 0.16 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr3_-_24909245 | 0.16 |
ENSDART00000178321
|
SHISA8
|
shisa family member 8 |
| chr14_-_46117345 | 0.16 |
ENSDART00000172166
|
NDUFC1
|
si:ch211-235e9.6 |
| chr11_-_16152400 | 0.16 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr7_+_11390462 | 0.15 |
ENSDART00000114383
|
tlnrd1
|
talin rod domain containing 1 |
| chr2_+_1988036 | 0.13 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr2_-_27774783 | 0.12 |
ENSDART00000161864
|
XKR4
|
zgc:123035 |
| chr9_+_32859967 | 0.11 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr20_+_36730049 | 0.10 |
ENSDART00000045948
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr20_+_39283849 | 0.08 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr12_+_15487834 | 0.04 |
ENSDART00000181138
|
arhgap23b
|
Rho GTPase activating protein 23b |
| chr21_-_40174647 | 0.04 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr7_-_71384391 | 0.02 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr20_-_8365244 | 0.01 |
ENSDART00000167102
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of bhlha15_bhlhe23+si:ch211-196c10.13_bhlhe22
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 3.8 | 15.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 2.0 | 6.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.0 | 5.9 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.8 | 13.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.6 | 8.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.6 | 3.1 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.6 | 6.6 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.6 | 7.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.6 | 12.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.5 | 2.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.5 | 2.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.5 | 10.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 4.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 1.9 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.3 | 9.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 5.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 2.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 2.6 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.3 | 0.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.3 | 14.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 1.2 | GO:0051103 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.2 | 3.4 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 5.0 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 0.9 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 11.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 0.6 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 1.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.8 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.9 | GO:1902837 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 1.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 2.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.7 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 0.9 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 10.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 3.0 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 1.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.7 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 1.4 | GO:1902850 | establishment of mitotic spindle localization(GO:0040001) mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 5.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 2.3 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 9.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.9 | 13.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 2.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.5 | 34.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.3 | 1.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 5.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 17.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 4.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 5.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 18.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 2.2 | 13.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 12.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.7 | 8.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.7 | 13.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 14.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.4 | 4.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 7.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 7.8 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 4.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 4.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 1.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.9 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 2.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 14.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 9.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 5.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 23.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 6.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 11.8 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 13.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 39.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.2 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.2 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 8.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 14.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 1.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 4.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 5.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 40.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 5.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 6.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |