Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
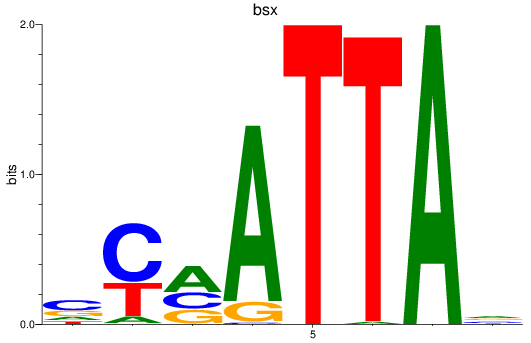
Results for bsx
Z-value: 0.51
Transcription factors associated with bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bsx
|
ENSDARG00000068976 | brain-specific homeobox |
|
bsx
|
ENSDARG00000110782 | brain-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bsx | dr11_v1_chr10_-_29892486_29892486 | -0.35 | 5.8e-04 | Click! |
Activity profile of bsx motif
Sorted Z-values of bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_29794058 | 3.91 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr11_+_18130300 | 3.73 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr20_-_23426339 | 3.62 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_-_10014512 | 3.26 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr10_-_25217347 | 3.00 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr21_+_28445052 | 3.00 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr11_+_18183220 | 2.98 |
ENSDART00000113468
|
LO018315.10
|
|
| chr11_+_18157260 | 2.95 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr21_+_25777425 | 2.83 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr11_-_1550709 | 2.82 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr12_-_5728755 | 2.81 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr8_-_554540 | 2.74 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr1_-_33645967 | 2.69 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr11_-_6452444 | 2.69 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr21_-_43666420 | 2.56 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr17_+_16046314 | 2.42 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_16046132 | 2.33 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr16_+_29509133 | 2.31 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr24_+_38306010 | 2.27 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr11_+_18175893 | 2.22 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr17_+_28102487 | 2.18 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr12_-_42368296 | 2.17 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr24_-_10021341 | 2.15 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr25_+_10410620 | 2.09 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr25_+_20089986 | 2.08 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr19_+_7567763 | 2.07 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr5_-_68333081 | 2.05 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr3_+_18398876 | 2.02 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr22_-_10586191 | 1.96 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr1_-_45049603 | 1.95 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr5_-_9216758 | 1.89 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr5_-_30615901 | 1.87 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_+_41588170 | 1.81 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr10_-_21362071 | 1.80 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_44560165 | 1.80 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr22_-_10121880 | 1.76 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_+_22851832 | 1.74 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr10_-_21362320 | 1.73 |
ENSDART00000189789
|
avd
|
avidin |
| chr22_-_10486477 | 1.72 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr24_-_25166720 | 1.71 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr15_-_23376541 | 1.69 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr3_-_53092509 | 1.68 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr25_-_29074064 | 1.62 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_-_33427803 | 1.61 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr16_-_17197546 | 1.61 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr18_+_7204378 | 1.60 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr10_-_34002185 | 1.60 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr13_-_50108337 | 1.59 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr11_+_24703108 | 1.59 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr22_-_37686966 | 1.58 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr21_-_3672343 | 1.57 |
ENSDART00000086492
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr2_-_38287987 | 1.56 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr16_+_50953547 | 1.56 |
ENSDART00000157526
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr19_-_20403507 | 1.52 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr3_-_53091946 | 1.51 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr12_+_22580579 | 1.50 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_31585917 | 1.48 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr4_+_14981854 | 1.48 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr9_-_6372535 | 1.46 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr16_-_45178430 | 1.44 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr22_+_22010128 | 1.41 |
ENSDART00000172610
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr11_-_7261717 | 1.40 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr22_+_35068046 | 1.40 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr3_+_23738215 | 1.39 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr3_-_3448095 | 1.38 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr24_+_12835935 | 1.36 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr1_-_513762 | 1.36 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr24_-_37640705 | 1.36 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr8_-_20243389 | 1.34 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr5_+_6670945 | 1.29 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr18_+_9171778 | 1.29 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr2_+_41926707 | 1.28 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr19_-_20403845 | 1.28 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr11_-_21031773 | 1.28 |
ENSDART00000065985
|
fmoda
|
fibromodulin a |
| chr12_-_35830625 | 1.28 |
ENSDART00000180028
|
CU459056.1
|
|
| chr6_-_46589726 | 1.26 |
ENSDART00000084334
|
ptgis
|
prostaglandin I2 (prostacyclin) synthase |
| chr11_+_5926850 | 1.26 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr3_+_30922947 | 1.25 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr24_+_34069675 | 1.24 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr23_-_19715557 | 1.23 |
ENSDART00000143764
|
rpl10
|
ribosomal protein L10 |
| chr20_-_22476255 | 1.22 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr5_+_27897504 | 1.21 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr2_+_20793982 | 1.21 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr10_-_35257458 | 1.20 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr6_+_28208973 | 1.20 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr7_+_17816006 | 1.17 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr1_+_45056371 | 1.17 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr1_-_55248496 | 1.15 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr22_-_15578402 | 1.13 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr21_+_20383837 | 1.12 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr8_-_23612462 | 1.12 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr13_-_40401870 | 1.11 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr23_-_31913069 | 1.11 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr6_+_39098397 | 1.10 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr2_+_33326522 | 1.10 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr6_-_19042294 | 1.10 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr21_-_22547496 | 1.10 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr3_-_26183699 | 1.09 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr20_+_26916639 | 1.09 |
ENSDART00000077787
|
serpinb1l2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 2 |
| chr2_-_51511494 | 1.09 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr20_-_40755614 | 1.08 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr18_-_18937485 | 1.08 |
ENSDART00000139015
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr18_+_48423973 | 1.08 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr22_-_10502780 | 1.07 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr19_-_12212692 | 1.06 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr11_+_37178271 | 1.06 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr24_-_4450238 | 1.05 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr14_-_21959712 | 1.05 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr15_-_31419805 | 1.04 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr25_+_3306620 | 1.03 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr3_-_16719244 | 1.03 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr24_-_25098719 | 1.03 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_+_40589770 | 1.03 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr11_+_27347076 | 1.03 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr1_-_43905252 | 1.02 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr21_-_7035599 | 1.01 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr19_-_25081711 | 1.01 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr4_-_2380173 | 1.00 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr11_-_28050559 | 1.00 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr15_+_1534644 | 1.00 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr21_-_2322102 | 1.00 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr9_+_24008879 | 0.99 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr23_+_39695827 | 0.97 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr23_+_38251864 | 0.97 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr13_+_17468161 | 0.97 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr10_-_41980797 | 0.97 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr20_-_42702832 | 0.96 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr3_-_32596394 | 0.95 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr17_+_6563307 | 0.94 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr24_-_41220538 | 0.92 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr23_-_31913231 | 0.91 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr19_+_19786117 | 0.90 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr25_+_13205878 | 0.90 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr16_+_33142734 | 0.90 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr6_-_43283122 | 0.90 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr2_-_20599315 | 0.90 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr6_+_41191482 | 0.90 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr18_+_50526290 | 0.90 |
ENSDART00000175194
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr17_-_10043273 | 0.89 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr3_+_6469754 | 0.89 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr23_+_36095260 | 0.88 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr25_+_3306858 | 0.88 |
ENSDART00000137077
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr17_+_30843881 | 0.88 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr3_+_26244353 | 0.88 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr19_-_10330778 | 0.87 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr6_-_35046735 | 0.87 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_+_23737795 | 0.86 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr7_-_38658411 | 0.86 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr14_-_33334065 | 0.85 |
ENSDART00000052761
|
rpl39
|
ribosomal protein L39 |
| chr10_+_6884627 | 0.85 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr10_+_6884123 | 0.84 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr3_-_19517462 | 0.84 |
ENSDART00000162027
|
CU571315.2
|
|
| chr23_-_5101847 | 0.84 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr1_-_59287410 | 0.83 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr21_-_11654422 | 0.82 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr5_+_11290851 | 0.82 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr12_-_33817114 | 0.82 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr20_+_20499869 | 0.81 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr2_-_37103622 | 0.81 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr24_-_2450597 | 0.81 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr14_+_22022441 | 0.80 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr5_+_63375386 | 0.80 |
ENSDART00000137855
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr4_-_9891874 | 0.80 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr7_+_17816470 | 0.79 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr7_-_64589920 | 0.79 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr19_+_4062101 | 0.76 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr15_+_41027466 | 0.76 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr25_+_34641536 | 0.76 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr16_+_33902006 | 0.76 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr12_-_48477031 | 0.75 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr19_-_20403318 | 0.75 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr17_-_6514962 | 0.74 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr24_+_39695688 | 0.73 |
ENSDART00000109747
|
zgc:153659
|
zgc:153659 |
| chr5_+_63375620 | 0.73 |
ENSDART00000185568
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr3_+_15828999 | 0.72 |
ENSDART00000104397
|
tmem11
|
transmembrane protein 11 |
| chr11_-_37997419 | 0.71 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr20_-_22378401 | 0.71 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr8_+_26253252 | 0.71 |
ENSDART00000142031
|
slc26a6
|
solute carrier family 26, member 6 |
| chr5_+_23242370 | 0.70 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr13_-_11984867 | 0.69 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr19_-_19871211 | 0.68 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr2_-_26596794 | 0.68 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr4_-_14191434 | 0.68 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr24_+_1023839 | 0.67 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr24_+_36636208 | 0.66 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr7_-_51368681 | 0.66 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr15_-_1822548 | 0.66 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr7_+_34592526 | 0.65 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr14_-_3381303 | 0.65 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr6_+_40922572 | 0.65 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr16_-_24661526 | 0.65 |
ENSDART00000058959
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr4_-_16853464 | 0.64 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr7_-_17591007 | 0.64 |
ENSDART00000171023
|
CU672228.1
|
|
| chr10_-_23099809 | 0.63 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr3_-_26184018 | 0.63 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr14_-_17622080 | 0.63 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr11_-_45138857 | 0.63 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr20_+_46513651 | 0.62 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr9_+_29548195 | 0.62 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr7_-_54679595 | 0.62 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.0 | 3.0 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.9 | 2.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 3.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.6 | 3.6 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.5 | 1.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 1.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.4 | 1.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.8 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 1.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.3 | 1.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 1.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 1.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 3.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.9 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.7 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.2 | 0.6 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 1.0 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.2 | 1.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.8 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 0.5 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.5 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 0.6 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 1.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.8 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.8 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 2.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.5 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.6 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.3 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 1.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 5.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.7 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.2 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.6 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.1 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 2.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 0.2 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.8 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.1 | GO:0022410 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.1 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 1.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 3.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0048897 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 3.4 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 1.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0045005 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.0 | 0.2 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 1.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.0 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.8 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.9 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 7.6 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 1.3 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.6 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.0 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.2 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.1 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 3.0 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.2 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 2.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 1.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 2.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 2.8 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 8.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 3.7 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.3 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 3.0 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 1.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 3.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 1.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.3 | 1.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.3 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 5.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.3 | 1.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 3.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.8 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 1.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.7 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.9 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.2 | 2.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 2.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 1.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 7.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 5.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 5.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.0 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |