Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
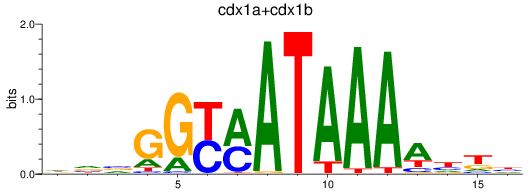
Results for cdx1a+cdx1b
Z-value: 0.70
Transcription factors associated with cdx1a+cdx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdx1a
|
ENSDARG00000114554 | caudal type homeobox 1a |
|
cdx1b
|
ENSDARG00000116139 | caudal type homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdx1b | dr11_v1_chr21_-_43992027_43992027 | -0.19 | 6.1e-02 | Click! |
| cdx1a | dr11_v1_chr14_+_3287740_3287740 | 0.01 | 9.1e-01 | Click! |
Activity profile of cdx1a+cdx1b motif
Sorted Z-values of cdx1a+cdx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_39105051 | 7.84 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr14_-_15171435 | 7.49 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr23_+_7379728 | 7.06 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr24_-_9997948 | 5.94 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_10014512 | 5.49 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr14_+_15495088 | 4.97 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr7_-_48263516 | 4.52 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr21_+_11969603 | 4.50 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr11_-_25257045 | 4.44 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr10_-_21545091 | 4.33 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr20_-_43741159 | 4.08 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr11_-_25257595 | 3.93 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr14_+_15597049 | 3.83 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr25_-_29074064 | 3.79 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr24_-_10006158 | 3.78 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr1_-_53468160 | 3.65 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr3_-_53465223 | 3.43 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr5_-_1963498 | 3.41 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr20_+_52546186 | 3.34 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_40321067 | 3.22 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr3_+_32526263 | 3.14 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr8_+_30709685 | 3.04 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr21_+_20396858 | 3.03 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr25_+_29161609 | 3.01 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr10_+_27068251 | 3.00 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr20_+_26880668 | 2.88 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr13_-_25719628 | 2.86 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr21_-_39024754 | 2.83 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr25_-_26758253 | 2.62 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr5_-_16351306 | 2.57 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr7_+_1473929 | 2.51 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr1_-_9249943 | 2.51 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr23_-_33750307 | 2.48 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr23_+_44349252 | 2.47 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr23_-_33750135 | 2.45 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr13_-_36034582 | 2.44 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr15_-_28596507 | 2.34 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr16_+_13993285 | 2.22 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr1_-_14332283 | 2.19 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr23_-_10722664 | 2.15 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr7_+_18075504 | 2.14 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr3_+_49074008 | 2.13 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr19_-_47571456 | 2.09 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_-_20840714 | 2.09 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr10_-_1625080 | 2.07 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr16_-_16225260 | 2.07 |
ENSDART00000165790
|
gra
|
granulito |
| chr10_+_44700103 | 2.06 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr7_+_66822229 | 2.05 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr8_-_38159805 | 2.01 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr15_+_22390076 | 1.99 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr5_+_64856666 | 1.99 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr11_+_24314148 | 1.98 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_-_6976096 | 1.97 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr17_-_30652738 | 1.91 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr7_-_60096318 | 1.88 |
ENSDART00000189125
|
BX511067.1
|
|
| chr11_-_25538341 | 1.86 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr14_+_34490445 | 1.81 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr8_-_39739627 | 1.77 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr12_+_20552190 | 1.75 |
ENSDART00000113224
|
gna13a
|
guanine nucleotide binding protein (G protein), alpha 13a |
| chr6_+_120181 | 1.74 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr14_-_3070613 | 1.74 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr11_+_20896122 | 1.73 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr12_-_6172154 | 1.72 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr9_+_32178374 | 1.72 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr2_-_40196547 | 1.71 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr14_-_4121052 | 1.71 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr16_+_20934353 | 1.69 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr24_-_36723116 | 1.67 |
ENSDART00000088204
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr19_+_3840955 | 1.65 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr7_-_55454406 | 1.64 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr21_-_2415808 | 1.62 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr14_-_4120636 | 1.61 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr8_-_39739056 | 1.59 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr2_-_17235891 | 1.59 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr6_+_39114345 | 1.58 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr16_+_52966812 | 1.58 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr5_+_38619813 | 1.55 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr3_+_36646054 | 1.53 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr22_+_34616151 | 1.51 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr14_+_32918172 | 1.51 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr25_+_20089986 | 1.50 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr24_+_5237753 | 1.48 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr19_+_19747430 | 1.47 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr23_-_3758637 | 1.46 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr11_-_22361306 | 1.45 |
ENSDART00000180688
ENSDART00000182200 ENSDART00000006580 |
tfeb
|
transcription factor EB |
| chr10_+_40629616 | 1.45 |
ENSDART00000147476
|
CR396590.9
|
|
| chr16_+_34046733 | 1.44 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| chr4_+_25630555 | 1.44 |
ENSDART00000133425
|
acot15
|
acyl-CoA thioesterase 15 |
| chr10_+_44699734 | 1.43 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr17_-_29192987 | 1.42 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr24_-_30275735 | 1.41 |
ENSDART00000168723
|
snx7
|
sorting nexin 7 |
| chr11_-_36051004 | 1.41 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr15_+_32405959 | 1.39 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr20_-_2641233 | 1.37 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr2_+_16798652 | 1.35 |
ENSDART00000145778
ENSDART00000087120 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr3_-_60589292 | 1.34 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr20_-_34670236 | 1.34 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr15_+_32423801 | 1.34 |
ENSDART00000165591
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr15_+_9297340 | 1.30 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr9_+_54290896 | 1.30 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr16_-_9675982 | 1.28 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr19_+_27970292 | 1.27 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr3_+_62161184 | 1.27 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr11_+_25257022 | 1.26 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr5_-_30984271 | 1.26 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr3_-_4501026 | 1.26 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr6_-_12588044 | 1.25 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr13_-_17860307 | 1.24 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr23_-_39751234 | 1.23 |
ENSDART00000160853
|
minos1
|
mitochondrial inner membrane organizing system 1 |
| chr2_+_52847049 | 1.23 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr20_-_29499363 | 1.22 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_9073433 | 1.22 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr11_+_12052791 | 1.19 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr10_+_26612321 | 1.17 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr11_-_15090118 | 1.17 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr2_+_16798923 | 1.16 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr23_+_36087219 | 1.16 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr18_+_14684115 | 1.14 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr16_+_54829574 | 1.13 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr12_+_34770531 | 1.13 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr8_-_6897619 | 1.13 |
ENSDART00000133606
|
si:ch211-255g12.8
|
si:ch211-255g12.8 |
| chr5_-_9625459 | 1.13 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr20_+_38458084 | 1.12 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr1_+_44780849 | 1.12 |
ENSDART00000145895
|
si:dkey-9i23.5
|
si:dkey-9i23.5 |
| chr2_-_5728843 | 1.11 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr2_+_19522082 | 1.11 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr10_-_15849027 | 1.10 |
ENSDART00000184682
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr5_+_38631119 | 1.10 |
ENSDART00000131832
ENSDART00000162215 |
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr23_-_43718067 | 1.08 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr15_-_22147860 | 1.08 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr23_+_43718115 | 1.07 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr5_-_57723929 | 1.06 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr5_-_23715861 | 1.05 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr9_-_22355391 | 1.05 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr9_+_3282369 | 1.04 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr2_+_22409249 | 1.03 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr14_+_35369979 | 1.00 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr12_-_33746111 | 1.00 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr1_-_13989643 | 1.00 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr9_+_2041535 | 0.99 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr19_+_1873059 | 0.99 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr6_+_39279831 | 0.97 |
ENSDART00000155088
|
ankrd33ab
|
ankyrin repeat domain 33Ab |
| chr2_-_6115389 | 0.97 |
ENSDART00000134921
|
prdx1
|
peroxiredoxin 1 |
| chr12_+_23912074 | 0.96 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr14_+_14841685 | 0.93 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr6_-_25165693 | 0.92 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr19_-_7225060 | 0.92 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr3_+_60589157 | 0.91 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr5_+_38679623 | 0.91 |
ENSDART00000131879
ENSDART00000170010 |
si:dkey-58f10.11
si:dkey-58f10.10
|
si:dkey-58f10.11 si:dkey-58f10.10 |
| chr20_+_7398090 | 0.90 |
ENSDART00000108649
|
pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr21_-_11970199 | 0.89 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr21_-_8153165 | 0.88 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr19_+_43822968 | 0.88 |
ENSDART00000171041
|
eloa
|
elongin A |
| chr11_-_15090564 | 0.88 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr18_+_17583479 | 0.87 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr15_+_37950556 | 0.86 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr1_+_34496855 | 0.86 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr1_-_7603734 | 0.86 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_+_8508753 | 0.86 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr20_-_49889111 | 0.84 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr24_+_30392834 | 0.84 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr18_-_15559817 | 0.81 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr14_+_3071110 | 0.80 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr3_-_53091946 | 0.80 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr10_+_35526528 | 0.80 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr8_-_25033681 | 0.79 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr9_-_34260214 | 0.79 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr2_-_19576640 | 0.78 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr9_-_710896 | 0.78 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr10_-_35410518 | 0.77 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr3_-_34624745 | 0.76 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr1_-_9486214 | 0.76 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr19_+_1872794 | 0.75 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr6_-_37745508 | 0.75 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr3_-_4557653 | 0.73 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr22_+_34615591 | 0.73 |
ENSDART00000189563
|
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr22_+_2512154 | 0.72 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr13_+_27329556 | 0.70 |
ENSDART00000140085
|
mb21d1
|
Mab-21 domain containing 1 |
| chr15_-_563877 | 0.70 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr2_-_19520690 | 0.70 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr12_-_2993095 | 0.69 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr21_-_41838284 | 0.69 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr2_+_14992879 | 0.68 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr3_-_31115601 | 0.67 |
ENSDART00000139090
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr2_+_35733335 | 0.66 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr12_-_16524727 | 0.66 |
ENSDART00000166645
|
FO704724.1
|
|
| chr2_-_19520324 | 0.65 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr1_-_43892349 | 0.65 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr6_-_39727244 | 0.64 |
ENSDART00000065057
|
itgb7
|
integrin, beta 7 |
| chr9_+_53637932 | 0.64 |
ENSDART00000188962
|
CABZ01084081.1
|
|
| chr25_+_17847054 | 0.63 |
ENSDART00000067290
|
PYURF
|
zgc:162634 |
| chr11_-_44622240 | 0.63 |
ENSDART00000172258
ENSDART00000173340 |
tbce
|
tubulin folding cofactor E |
| chr13_+_13668991 | 0.63 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr2_+_19578446 | 0.60 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_-_9744081 | 0.60 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr18_+_9171778 | 0.60 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr13_-_5252559 | 0.60 |
ENSDART00000181652
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr1_-_43897831 | 0.59 |
ENSDART00000048225
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr15_-_4580763 | 0.59 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_-_12713920 | 0.58 |
ENSDART00000099749
|
CLDN22
|
si:dkey-98f17.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdx1a+cdx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 1.2 | 3.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.9 | 4.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.8 | 4.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.7 | 2.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.7 | 2.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.7 | 2.1 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.7 | 2.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.6 | 3.0 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.6 | 1.8 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.5 | 3.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.4 | 1.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 1.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.4 | 0.8 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.4 | 8.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.4 | 2.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 2.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 2.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.3 | 1.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 1.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 1.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 3.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.3 | 3.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.3 | 1.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 1.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 2.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 0.9 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 1.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 1.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 3.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.6 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 3.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.4 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.7 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.7 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 1.0 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.9 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.6 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.6 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 3.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 2.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 3.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 1.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 3.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.7 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 2.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 3.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 1.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.0 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 1.8 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 2.5 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 3.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.4 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.2 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 2.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 1.2 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 5.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.6 | 1.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.5 | 2.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 2.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 3.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 2.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 3.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 2.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 3.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 3.3 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 2.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.7 | 2.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.7 | 3.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.6 | 3.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.6 | 1.8 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.6 | 3.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.5 | 3.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 3.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 2.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 0.8 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.2 | 0.9 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.2 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.8 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 1.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.8 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 1.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 2.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 4.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 4.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.4 | 3.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 3.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 8.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |