Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
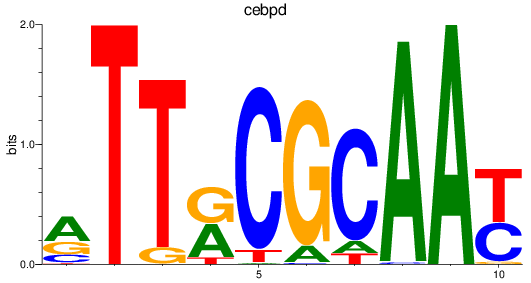
Results for cebpd
Z-value: 1.16
Transcription factors associated with cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpd
|
ENSDARG00000087303 | CCAAT enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpd | dr11_v1_chr24_+_35564668_35564668 | 0.05 | 6.1e-01 | Click! |
Activity profile of cebpd motif
Sorted Z-values of cebpd motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_63509581 | 24.84 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr1_-_10071422 | 22.85 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr20_+_23440632 | 22.11 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr8_-_39739627 | 20.95 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr16_+_23984755 | 16.88 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr14_+_16345003 | 16.83 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr4_-_17409533 | 13.96 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr16_-_27749172 | 13.23 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr20_-_40755614 | 12.01 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr20_-_40750953 | 11.42 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr16_+_44298902 | 11.17 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr21_+_17051478 | 11.01 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr22_-_26175237 | 10.41 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr14_+_21106444 | 9.17 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr14_+_21107032 | 9.02 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr5_-_69948099 | 8.79 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr16_+_50289916 | 8.72 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr22_-_17595310 | 8.70 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr16_+_23984179 | 8.66 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr6_+_10333920 | 7.87 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr24_-_32665283 | 7.73 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr9_-_9225980 | 7.56 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr4_-_17391091 | 7.19 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr7_+_67733759 | 6.94 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr15_-_29348212 | 6.25 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr8_+_17884569 | 5.93 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr21_-_39058490 | 5.55 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr17_-_6076084 | 5.51 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_+_30699429 | 5.51 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr2_-_51772438 | 5.34 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr17_-_6076266 | 5.24 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr9_+_38292947 | 5.04 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr22_+_15336752 | 4.89 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr12_+_4920451 | 4.86 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr2_-_37277626 | 4.80 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr6_+_9130989 | 4.67 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr8_+_42917515 | 4.63 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr9_-_53062083 | 4.38 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr20_+_25563105 | 4.29 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr5_+_8196264 | 4.25 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr25_-_12805295 | 4.21 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr5_+_25585869 | 3.84 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr7_-_7823662 | 3.75 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr12_+_46960579 | 3.72 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr20_-_19511700 | 3.71 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr17_+_10318071 | 3.66 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr12_+_25775734 | 3.56 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr5_-_43935460 | 3.38 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr5_+_51111343 | 3.18 |
ENSDART00000092002
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr15_-_17869115 | 3.14 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr3_+_36972298 | 3.02 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr15_-_17868870 | 2.99 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr12_-_22238004 | 2.90 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr15_-_17960228 | 2.90 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr16_-_17072440 | 2.89 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr10_+_38526496 | 2.87 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr5_+_51111766 | 2.78 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr7_-_39751540 | 2.74 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr12_-_17592215 | 2.74 |
ENSDART00000134597
|
usp42
|
ubiquitin specific peptidase 42 |
| chr9_+_2574122 | 2.63 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr3_+_19216567 | 2.63 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr24_-_36727922 | 2.60 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr11_+_43419809 | 2.58 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr5_-_54395488 | 2.50 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr9_+_9502610 | 2.41 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr19_+_1370504 | 2.37 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr7_-_46019756 | 2.28 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr6_+_44197348 | 2.24 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr17_+_18031899 | 2.20 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr14_-_38929885 | 2.20 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr5_-_43935119 | 2.17 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr7_-_52963493 | 2.07 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr21_-_14762944 | 2.05 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr19_+_46259619 | 2.04 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr17_-_10838434 | 2.03 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr9_-_7238839 | 2.01 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr13_-_30996072 | 1.97 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr7_+_15736230 | 1.94 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr8_+_23703464 | 1.94 |
ENSDART00000132584
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr12_-_1085227 | 1.93 |
ENSDART00000169936
|
sox8a
|
SRY (sex determining region Y)-box 8a |
| chr7_+_65240227 | 1.91 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr6_-_39649504 | 1.90 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr1_+_14253118 | 1.87 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr14_-_38809561 | 1.83 |
ENSDART00000159159
|
sil1
|
SIL1 nucleotide exchange factor |
| chr3_+_36972586 | 1.81 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr20_+_37820992 | 1.79 |
ENSDART00000064692
|
tatdn3
|
TatD DNase domain containing 3 |
| chr20_-_33675676 | 1.78 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr6_+_44197099 | 1.76 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr11_-_6265574 | 1.63 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr19_+_8606883 | 1.60 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr13_-_25548733 | 1.60 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr22_-_16377960 | 1.59 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr20_-_45661049 | 1.56 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr6_+_11397269 | 1.54 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr19_-_20446756 | 1.54 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr11_+_13176568 | 1.54 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr2_-_10098191 | 1.53 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr7_+_38089650 | 1.50 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr7_-_37895771 | 1.48 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr24_-_39772045 | 1.48 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr14_+_7902374 | 1.48 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr14_-_8890437 | 1.47 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr3_-_30152836 | 1.45 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr5_-_23574234 | 1.44 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_7246848 | 1.42 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr23_+_44732863 | 1.36 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr25_+_3294150 | 1.34 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr19_-_15192638 | 1.29 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr8_+_4368534 | 1.26 |
ENSDART00000015214
|
pptc7a
|
PTC7 protein phosphatase homolog a |
| chr3_-_30153242 | 1.24 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr24_-_31123365 | 1.23 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr22_+_10158502 | 1.23 |
ENSDART00000005869
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr12_+_13205955 | 1.23 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr18_-_16922905 | 1.22 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr15_+_20543770 | 1.21 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr12_+_17154655 | 1.18 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr5_-_45894802 | 1.17 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr5_-_26893310 | 1.15 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr1_+_26071869 | 1.15 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr20_-_27381691 | 1.13 |
ENSDART00000010293
|
serpina10a
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a |
| chr7_-_22790630 | 1.13 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr7_+_23580082 | 1.13 |
ENSDART00000183245
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr22_+_15973122 | 1.10 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr13_-_32898962 | 1.06 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr16_-_7793457 | 1.03 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr11_+_39135050 | 1.02 |
ENSDART00000180571
ENSDART00000189685 |
cdc42
|
cell division cycle 42 |
| chr5_-_48680580 | 0.99 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr7_-_7845540 | 0.93 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr16_+_14029283 | 0.92 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr20_-_25533739 | 0.89 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr9_-_12034444 | 0.83 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr14_-_5642371 | 0.83 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr13_-_18195942 | 0.79 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr4_-_14207471 | 0.77 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr17_-_20236228 | 0.77 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr18_+_507835 | 0.77 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr8_-_23702295 | 0.76 |
ENSDART00000162296
|
cfp
|
complement factor properdin |
| chr2_-_36819624 | 0.74 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr17_-_42799104 | 0.74 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr12_-_13205572 | 0.74 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr3_+_41731527 | 0.72 |
ENSDART00000049007
ENSDART00000187866 |
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr19_+_16064439 | 0.66 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr8_+_12930216 | 0.65 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr22_-_10158038 | 0.65 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr10_-_29733194 | 0.64 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr3_-_43821191 | 0.63 |
ENSDART00000186816
|
snn
|
stannin |
| chr20_-_33961697 | 0.61 |
ENSDART00000061765
|
selp
|
selectin P |
| chr12_-_13205854 | 0.61 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr16_+_46111849 | 0.60 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr17_+_23975762 | 0.60 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr2_-_24348948 | 0.60 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr21_-_43550120 | 0.59 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr17_+_43013171 | 0.59 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr13_-_25842074 | 0.58 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr3_-_31079186 | 0.52 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr17_+_28670132 | 0.46 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr9_-_32300611 | 0.46 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr4_-_5302866 | 0.39 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr8_-_23701880 | 0.38 |
ENSDART00000139897
|
si:ch73-237c6.1
|
si:ch73-237c6.1 |
| chr2_+_11029138 | 0.37 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr5_-_29488245 | 0.37 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr2_-_50298337 | 0.35 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr11_+_23933016 | 0.35 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr6_+_8314451 | 0.35 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr2_-_4032732 | 0.34 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr16_-_27138478 | 0.34 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr22_+_24645325 | 0.34 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr5_+_32162684 | 0.33 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr7_+_12950507 | 0.29 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr10_-_20523405 | 0.29 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr18_-_46369516 | 0.29 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr5_-_43682930 | 0.28 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr4_-_5302162 | 0.23 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr24_-_15648636 | 0.22 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr4_+_21867522 | 0.21 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr24_+_11083146 | 0.17 |
ENSDART00000009473
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr19_-_9829965 | 0.17 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr5_-_69437422 | 0.16 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr18_+_16330025 | 0.15 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr9_-_35069645 | 0.13 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr6_-_11780070 | 0.12 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr20_+_2950005 | 0.12 |
ENSDART00000135919
|
amd1
|
adenosylmethionine decarboxylase 1 |
| chr24_+_119680 | 0.12 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr3_-_46818001 | 0.12 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr24_+_38671054 | 0.10 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr23_-_36003282 | 0.08 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr9_-_32300783 | 0.06 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr13_+_40437550 | 0.02 |
ENSDART00000057090
ENSDART00000167859 |
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr2_-_30460293 | 0.02 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr8_-_31716302 | 0.02 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr4_+_71989418 | 0.00 |
ENSDART00000170996
|
parp11
|
poly(ADP-ribose) polymerase family member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpd
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0015677 | copper ion import(GO:0015677) |
| 3.1 | 9.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 2.5 | 7.6 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 2.4 | 7.2 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 1.9 | 7.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.6 | 4.8 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 1.6 | 6.3 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 1.5 | 22.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 1.3 | 25.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 1.2 | 8.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 1.2 | 14.0 | GO:0006570 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) tyrosine metabolic process(GO:0006570) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 1.1 | 5.5 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.8 | 18.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 11.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.7 | 2.9 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.7 | 4.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 1.6 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.4 | 2.6 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.4 | 3.7 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 2.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.4 | 10.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) venous blood vessel development(GO:0060841) |
| 0.4 | 1.6 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 6.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.4 | 3.7 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.4 | 1.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.3 | 1.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 4.7 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.3 | 5.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.3 | 2.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 2.8 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.2 | 2.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.9 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 1.9 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 1.0 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 1.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 4.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.4 | GO:0008343 | adult feeding behavior(GO:0008343) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 2.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 3.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 4.2 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.1 | 1.8 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 5.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 5.5 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 7.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 1.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 2.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.8 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 1.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.8 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.2 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 4.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.7 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 2.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.6 | 25.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.3 | 4.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.7 | 2.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.7 | 2.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 9.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 1.6 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 7.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 10.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 13.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 2.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 13.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 36.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 8.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 3.3 | 13.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.3 | 21.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 2.3 | 18.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.2 | 8.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.7 | 8.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.6 | 6.6 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.5 | 7.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.9 | 11.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.9 | 4.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.7 | 16.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.6 | 11.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 2.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.6 | 5.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.5 | 2.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 11.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 6.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 2.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 2.0 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.4 | 2.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 2.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 2.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 1.9 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 11.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 33.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.2 | 5.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 3.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 6.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 5.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 5.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 26.1 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 2.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 3.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 4.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 7.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.1 | GO:0004518 | nuclease activity(GO:0004518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 22.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 21.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 2.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 25.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.1 | 16.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.9 | 22.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.0 | 11.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.9 | 23.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.9 | 18.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 3.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 2.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 14.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |