Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
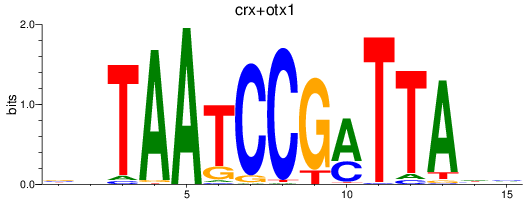
Results for crx+otx1
Z-value: 0.74
Transcription factors associated with crx+otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crx
|
ENSDARG00000011989 | cone-rod homeobox |
|
otx1
|
ENSDARG00000094992 | orthodenticle homeobox 1 |
|
crx
|
ENSDARG00000113850 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx1 | dr11_v1_chr17_+_24318753_24318753 | 0.67 | 2.4e-13 | Click! |
| crx | dr11_v1_chr5_+_36932718_36932718 | 0.25 | 1.6e-02 | Click! |
Activity profile of crx+otx1 motif
Sorted Z-values of crx+otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_38079261 | 7.05 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr19_-_5207361 | 6.80 |
ENSDART00000174611
|
stx1b
|
syntaxin 1B |
| chr24_-_21689146 | 6.04 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr21_+_13366353 | 5.54 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr25_+_21324588 | 5.52 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr3_+_62000822 | 5.51 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr11_-_3366782 | 4.83 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr3_-_13068189 | 4.62 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr10_+_6013076 | 4.51 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr13_+_733027 | 4.38 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr19_+_14921000 | 4.35 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr9_-_38042823 | 4.09 |
ENSDART00000141998
ENSDART00000133998 |
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr10_+_17026870 | 4.04 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr11_-_37693019 | 3.82 |
ENSDART00000102898
|
zgc:158258
|
zgc:158258 |
| chr8_-_46572298 | 3.64 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr16_+_43219363 | 3.63 |
ENSDART00000183610
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr16_-_46579936 | 3.57 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr2_+_20430366 | 3.42 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr24_-_31140356 | 3.34 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr4_+_72797711 | 3.30 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr18_+_27439680 | 3.28 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr25_-_19090479 | 3.15 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr21_-_28340977 | 3.11 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr22_+_17203752 | 3.05 |
ENSDART00000143376
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr6_-_58757131 | 2.93 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr13_-_29406534 | 2.85 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr23_+_2560005 | 2.83 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr10_-_42898220 | 2.72 |
ENSDART00000099270
|
CU326366.2
|
|
| chr8_-_4618653 | 2.70 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr20_-_10120442 | 2.68 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr10_+_40235959 | 2.66 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr14_-_4273396 | 2.50 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr6_-_10168822 | 2.38 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr2_-_39036604 | 2.36 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr21_-_24632778 | 2.32 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr1_+_35949269 | 2.26 |
ENSDART00000180628
|
BX276105.3
|
|
| chr17_-_7371564 | 2.25 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr15_+_46605482 | 2.24 |
ENSDART00000184449
|
CABZ01044048.1
|
|
| chr22_+_25453334 | 2.21 |
ENSDART00000123962
|
si:ch211-12h2.6
|
si:ch211-12h2.6 |
| chr16_+_9726038 | 2.19 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr3_+_15296824 | 2.15 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr7_+_67486807 | 2.12 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr23_+_7710721 | 2.09 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr13_-_40499296 | 2.08 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr11_-_1550709 | 2.08 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr13_-_30161684 | 2.07 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr21_-_39058490 | 2.03 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr21_+_33503835 | 2.00 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr20_+_13969414 | 1.98 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr21_+_31838386 | 1.97 |
ENSDART00000135591
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr2_+_44972720 | 1.94 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr5_-_27993972 | 1.90 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr19_+_45970692 | 1.87 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr19_-_32641725 | 1.82 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr6_-_16406210 | 1.77 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr5_+_66355153 | 1.76 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr23_+_45512825 | 1.76 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr20_-_54014373 | 1.73 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr9_+_21151138 | 1.72 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr4_+_11053301 | 1.70 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr18_-_8579907 | 1.70 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr25_-_204019 | 1.70 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr11_+_19080400 | 1.69 |
ENSDART00000044423
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr2_-_42864472 | 1.69 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr10_-_40968095 | 1.66 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr25_+_35913614 | 1.66 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr13_+_15581270 | 1.63 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr2_-_32387441 | 1.63 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr20_-_54014539 | 1.62 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr14_+_52413846 | 1.60 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr2_-_15349382 | 1.58 |
ENSDART00000057238
|
olfm3b
|
olfactomedin 3b |
| chr20_-_20932760 | 1.54 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_+_12111154 | 1.54 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr9_+_17982737 | 1.53 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr9_+_10014817 | 1.52 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr16_-_16701718 | 1.48 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr2_+_33052169 | 1.46 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr7_+_39011355 | 1.45 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr18_-_10713414 | 1.42 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr3_-_15679107 | 1.40 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr3_-_20058417 | 1.37 |
ENSDART00000165695
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr2_-_31686353 | 1.36 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr25_+_18964782 | 1.34 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr3_+_32842825 | 1.34 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr23_+_16935494 | 1.32 |
ENSDART00000143120
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr3_-_12903609 | 1.31 |
ENSDART00000160522
|
ccdc137
|
coiled-coil domain containing 137 |
| chr19_+_43884120 | 1.30 |
ENSDART00000139684
ENSDART00000142312 |
lypla2
|
lysophospholipase II |
| chr14_-_25985698 | 1.30 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr16_+_38159758 | 1.29 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr14_-_8816933 | 1.28 |
ENSDART00000163383
|
si:dkeyp-115e12.7
|
si:dkeyp-115e12.7 |
| chr5_-_27994679 | 1.25 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr4_+_5196469 | 1.23 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr10_+_39476432 | 1.23 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr10_-_25852517 | 1.22 |
ENSDART00000191551
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr2_+_49417900 | 1.21 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr22_-_16154771 | 1.20 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr16_+_4055331 | 1.19 |
ENSDART00000128978
|
CR391998.1
|
|
| chr1_-_17650223 | 1.18 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr3_-_15210491 | 1.17 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr17_+_18031899 | 1.15 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr21_+_36581384 | 1.14 |
ENSDART00000183924
|
BX936439.1
|
|
| chr17_+_2130018 | 1.14 |
ENSDART00000193675
ENSDART00000110529 |
bub1bb
|
BUB1 mitotic checkpoint serine/threonine kinase Bb |
| chr14_+_11909966 | 1.14 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr17_+_14886828 | 1.12 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr12_-_35883814 | 1.12 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr2_+_24722771 | 1.12 |
ENSDART00000147207
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr25_+_32031536 | 1.12 |
ENSDART00000173656
|
tjp1b
|
tight junction protein 1b |
| chr19_-_3106447 | 1.12 |
ENSDART00000137987
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr13_+_41917606 | 1.09 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr2_-_32551178 | 1.09 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr7_+_39011852 | 1.08 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr24_+_10039165 | 1.08 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr18_+_10784730 | 1.08 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr16_-_16225260 | 1.07 |
ENSDART00000165790
|
gra
|
granulito |
| chr9_+_10014514 | 1.07 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr7_-_39378903 | 1.07 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr18_-_24989580 | 1.06 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr15_-_35246742 | 1.06 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr8_-_19649617 | 1.05 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr11_+_18873113 | 1.05 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr14_-_6402769 | 1.03 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr11_-_41996957 | 1.03 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr1_+_28031697 | 1.03 |
ENSDART00000047211
ENSDART00000149785 ENSDART00000149817 ENSDART00000149479 ENSDART00000187922 |
chst10
|
carbohydrate sulfotransferase 10 |
| chr25_+_7532627 | 1.02 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr17_+_10593398 | 1.01 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr6_-_18121075 | 1.01 |
ENSDART00000171072
|
SEC14L1
|
si:dkey-237i9.1 |
| chr7_-_30492261 | 1.01 |
ENSDART00000173954
|
adam10a
|
ADAM metallopeptidase domain 10a |
| chr3_+_22327738 | 1.01 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr12_+_49100365 | 0.98 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr6_-_39218609 | 0.95 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr5_+_26847190 | 0.95 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr8_-_45430817 | 0.92 |
ENSDART00000150067
ENSDART00000112394 |
ywhabb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b |
| chr21_-_44104600 | 0.91 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr13_-_36844945 | 0.89 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr21_-_40317035 | 0.89 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr24_-_8730913 | 0.86 |
ENSDART00000187228
ENSDART00000082349 ENSDART00000186660 |
tfap2a
|
transcription factor AP-2 alpha |
| chr3_+_34140507 | 0.84 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr23_+_5465806 | 0.84 |
ENSDART00000149434
ENSDART00000148506 |
tulp1a
|
tubby like protein 1a |
| chr21_-_40562705 | 0.81 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr8_+_18545933 | 0.80 |
ENSDART00000148806
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr11_+_29770966 | 0.80 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr18_+_3634652 | 0.80 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr6_-_1780633 | 0.79 |
ENSDART00000160670
|
ntan1
|
N-terminal asparagine amidase |
| chr20_+_18260358 | 0.79 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_+_37623693 | 0.79 |
ENSDART00000144812
ENSDART00000182709 |
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr1_-_45580787 | 0.79 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_58766424 | 0.79 |
ENSDART00000191010
|
CABZ01096573.1
|
|
| chr23_+_21492151 | 0.79 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr16_+_33987892 | 0.78 |
ENSDART00000166302
|
pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr4_-_908821 | 0.78 |
ENSDART00000168266
|
mrap2b
|
melanocortin 2 receptor accessory protein 2b |
| chr10_+_26652859 | 0.77 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr10_+_4875262 | 0.76 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr11_-_27702778 | 0.75 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr22_+_5176255 | 0.75 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr14_+_11289851 | 0.74 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr12_-_33659328 | 0.74 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr5_-_40910749 | 0.73 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr9_+_3519191 | 0.71 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr8_+_31435452 | 0.70 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr22_+_2431585 | 0.70 |
ENSDART00000167758
|
zgc:171435
|
zgc:171435 |
| chr5_+_61799629 | 0.70 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr21_-_2209012 | 0.70 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr9_-_8314028 | 0.67 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr12_-_9084640 | 0.66 |
ENSDART00000125230
|
exoc6
|
exocyst complex component 6 |
| chr5_+_51833305 | 0.65 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr1_+_44523516 | 0.64 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr14_+_21754521 | 0.62 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr15_-_26930999 | 0.62 |
ENSDART00000181674
ENSDART00000126046 |
ccdc9
|
coiled-coil domain containing 9 |
| chr25_+_29694158 | 0.61 |
ENSDART00000174568
|
brd1b
|
bromodomain containing 1b |
| chr5_-_65081600 | 0.61 |
ENSDART00000160850
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr21_+_11367253 | 0.61 |
ENSDART00000091835
|
gtf3c5
|
general transcription factor IIIC, polypeptide 5 |
| chr7_-_26518086 | 0.60 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr14_+_14027427 | 0.58 |
ENSDART00000183204
|
rraga
|
Ras-related GTP binding A |
| chr18_+_19006063 | 0.58 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr6_+_54538948 | 0.58 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr3_+_22999849 | 0.56 |
ENSDART00000127239
|
si:ch211-237i5.4
|
si:ch211-237i5.4 |
| chr2_-_51757328 | 0.56 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr8_+_53120278 | 0.55 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr8_+_2456854 | 0.54 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr11_+_6295370 | 0.53 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr25_-_10629940 | 0.53 |
ENSDART00000154483
ENSDART00000155231 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr5_+_9434288 | 0.52 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr9_+_19095023 | 0.52 |
ENSDART00000110457
ENSDART00000099426 ENSDART00000137087 |
crfb1
|
cytokine receptor family member b1 |
| chr22_-_38934989 | 0.51 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr13_+_31002658 | 0.50 |
ENSDART00000140257
|
lrrc18a
|
leucine rich repeat containing 18a |
| chr4_-_18455480 | 0.50 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr5_-_32489796 | 0.48 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr3_+_50602866 | 0.46 |
ENSDART00000190069
ENSDART00000153921 |
gsg1l2a
|
gsg1-like 2a |
| chr22_+_5176693 | 0.46 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr20_+_42537768 | 0.45 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr10_-_41400049 | 0.45 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr4_-_56083825 | 0.44 |
ENSDART00000157710
|
znf986
|
zinc finger protein 986 |
| chr16_+_54592907 | 0.44 |
ENSDART00000172113
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr4_-_28353538 | 0.44 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr22_-_619711 | 0.41 |
ENSDART00000192778
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr20_-_45423498 | 0.40 |
ENSDART00000098424
|
trib2
|
tribbles pseudokinase 2 |
| chr23_+_30827959 | 0.39 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr2_+_30721070 | 0.38 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr7_+_20030888 | 0.38 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr5_-_23715861 | 0.38 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of crx+otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 1.5 | 4.5 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 1.4 | 6.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.1 | 4.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.3 | 2.9 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.3 | 4.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.3 | 2.1 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 0.7 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 1.8 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 1.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 3.6 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.2 | 1.0 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.2 | 3.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.6 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 1.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.5 | GO:0032241 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 2.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 4.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 1.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 1.8 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 1.7 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 5.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 2.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.8 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.8 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.9 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.3 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.8 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 2.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.0 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.3 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 3.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 1.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 2.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 4.8 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 0.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.7 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 1.0 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.2 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.2 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 4.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 0.9 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.3 | 6.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.8 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.5 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 2.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 6.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 4.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 6.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.4 | 4.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 1.0 | 4.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.7 | 2.9 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.7 | 4.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 3.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 2.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 1.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 3.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.3 | 4.8 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.3 | 1.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 2.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.8 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.2 | 2.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 6.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 6.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 3.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 5.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 6.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 6.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 4.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |