Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
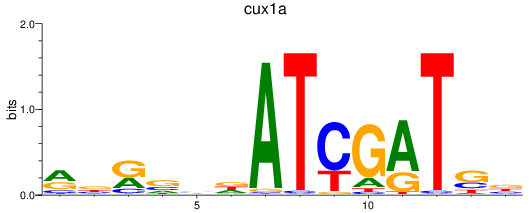
Results for cux1a
Z-value: 0.33
Transcription factors associated with cux1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux1a
|
ENSDARG00000078459 | cut-like homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux1a | dr11_v1_chr10_-_33156789_33156789 | -0.21 | 3.9e-02 | Click! |
Activity profile of cux1a motif
Sorted Z-values of cux1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_1625976 | 2.02 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr18_-_40481028 | 1.52 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr1_-_49932040 | 1.51 |
ENSDART00000048281
|
cyp2u1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr25_+_21324588 | 1.47 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr3_-_38692920 | 1.23 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr10_+_34377697 | 1.16 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr16_+_25126935 | 1.15 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr20_-_18736281 | 1.12 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr17_+_12159529 | 1.10 |
ENSDART00000046802
|
sccpdha
|
saccharopine dehydrogenase a |
| chr16_+_39242339 | 0.97 |
ENSDART00000102510
|
zgc:77056
|
zgc:77056 |
| chr15_-_9419472 | 0.97 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr23_-_33986762 | 0.94 |
ENSDART00000144609
|
tmed4
|
transmembrane p24 trafficking protein 4 |
| chr10_+_21786656 | 0.93 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr9_-_35334642 | 0.91 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr23_-_24047054 | 0.89 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr17_+_45454943 | 0.84 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr6_-_29105727 | 0.81 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr14_-_33083539 | 0.81 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr12_+_10115964 | 0.75 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr15_-_7598294 | 0.74 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr14_-_25985698 | 0.74 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr8_-_24135171 | 0.73 |
ENSDART00000112926
|
adora1b
|
adenosine A1 receptor b |
| chr15_-_7598542 | 0.73 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr4_-_25485404 | 0.69 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr2_+_23007675 | 0.65 |
ENSDART00000163649
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr11_+_41089574 | 0.65 |
ENSDART00000170023
|
phf13
|
PHD finger protein 13 |
| chr12_-_7607114 | 0.63 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr17_+_37249736 | 0.61 |
ENSDART00000189686
|
CR318625.1
|
|
| chr19_+_14059349 | 0.61 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr16_-_25380903 | 0.60 |
ENSDART00000086375
ENSDART00000188587 |
adnp2a
|
ADNP homeobox 2a |
| chr17_+_24597001 | 0.59 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr7_-_25895189 | 0.56 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr23_+_9353552 | 0.56 |
ENSDART00000163298
|
BX511246.1
|
|
| chr4_-_52189449 | 0.56 |
ENSDART00000192426
|
BX649431.4
|
|
| chr23_-_44494401 | 0.54 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr6_+_38896158 | 0.53 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr7_-_39751540 | 0.53 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr7_+_33152723 | 0.52 |
ENSDART00000132658
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr25_-_6082509 | 0.50 |
ENSDART00000104755
|
cpeb1a
|
cytoplasmic polyadenylation element binding protein 1a |
| chr21_-_19919918 | 0.48 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr2_-_32513538 | 0.47 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr8_-_29851706 | 0.46 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_-_39976492 | 0.44 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr5_-_13835461 | 0.44 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr12_-_9700605 | 0.43 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr11_-_20956309 | 0.43 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr5_-_26893310 | 0.41 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr11_+_16152316 | 0.40 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr3_-_30152836 | 0.37 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr15_-_16704417 | 0.37 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr11_-_12705608 | 0.37 |
ENSDART00000158937
ENSDART00000114391 |
zgc:174353
|
zgc:174353 |
| chr7_+_29951997 | 0.36 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr3_-_30153242 | 0.36 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr14_-_32486757 | 0.35 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr10_-_11397590 | 0.35 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr4_-_51460013 | 0.32 |
ENSDART00000193382
|
CR628395.1
|
|
| chr17_+_11507131 | 0.31 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr11_-_12673637 | 0.31 |
ENSDART00000186058
|
CR450764.7
|
|
| chr22_+_8012532 | 0.30 |
ENSDART00000159439
|
CABZ01034698.1
|
|
| chr12_-_19091214 | 0.28 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr13_-_36264861 | 0.28 |
ENSDART00000100204
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr16_+_4658250 | 0.27 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr24_+_35881124 | 0.26 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr22_-_15562933 | 0.24 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr12_+_38770654 | 0.24 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr2_-_37353098 | 0.23 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr16_-_12095144 | 0.21 |
ENSDART00000145106
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr12_+_14149686 | 0.20 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr13_-_4664403 | 0.17 |
ENSDART00000023803
ENSDART00000177957 |
c1d
|
C1D nuclear receptor corepressor |
| chr24_+_31374324 | 0.16 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr3_-_31079186 | 0.15 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr7_-_56766973 | 0.13 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr8_-_19451487 | 0.13 |
ENSDART00000184037
|
zgc:92140
|
zgc:92140 |
| chr22_+_29067388 | 0.12 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr20_+_32406011 | 0.12 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr2_+_17451656 | 0.10 |
ENSDART00000163620
|
BX640408.1
|
|
| chr10_+_44057177 | 0.10 |
ENSDART00000164610
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr5_+_37729207 | 0.10 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr2_-_51275873 | 0.09 |
ENSDART00000168019
|
si:ch211-215e19.4
|
si:ch211-215e19.4 |
| chr2_+_7715810 | 0.08 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr11_-_16152105 | 0.08 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr17_+_48164536 | 0.07 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr16_+_19014886 | 0.07 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr22_+_21516689 | 0.06 |
ENSDART00000105550
ENSDART00000136374 |
mier2
|
mesoderm induction early response 1, family member 2 |
| chr11_-_16152400 | 0.05 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr17_-_30839338 | 0.05 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr7_-_12789251 | 0.04 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr11_+_1522136 | 0.03 |
ENSDART00000002318
|
srsf6b
|
serine/arginine-rich splicing factor 6b |
| chr5_+_41996889 | 0.02 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr18_+_45571378 | 0.01 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr7_+_49272702 | 0.01 |
ENSDART00000083389
|
abtb2a
|
ankyrin repeat and BTB (POZ) domain containing 2a |
| chr2_-_37352514 | 0.01 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr7_+_29863548 | 0.00 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.5 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.3 | 0.8 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.3 | 1.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |