Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
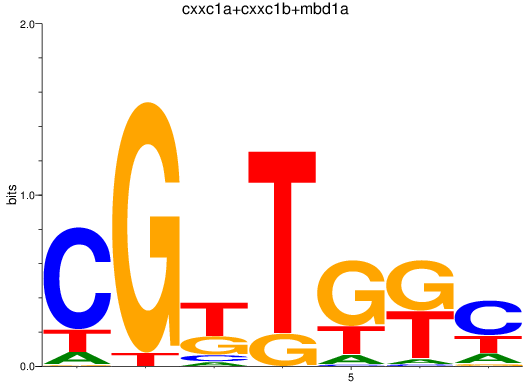
Results for cxxc1a+cxxc1b+mbd1a
Z-value: 0.14
Transcription factors associated with cxxc1a+cxxc1b+mbd1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cxxc1b
|
ENSDARG00000025718 | CXXC finger protein 1b |
|
mbd1a
|
ENSDARG00000101205 | methyl-CpG binding domain protein 1a |
|
cxxc1a
|
ENSDARG00000101996 | CXXC finger protein 1a |
|
cxxc1b
|
ENSDARG00000111907 | CXXC finger protein 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cxxc1a | dr11_v1_chr8_+_7854130_7854130 | -0.43 | 1.7e-05 | Click! |
| cxxc1b | dr11_v1_chr11_+_25539698_25539698 | -0.37 | 2.5e-04 | Click! |
| mbd1a | dr11_v1_chr8_+_7875110_7875142 | -0.37 | 2.8e-04 | Click! |
Activity profile of cxxc1a+cxxc1b+mbd1a motif
Sorted Z-values of cxxc1a+cxxc1b+mbd1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_38509333 | 0.61 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr23_-_20345473 | 0.46 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr21_+_27448856 | 0.46 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr11_+_44579865 | 0.43 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr21_-_28901095 | 0.38 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr22_-_6988102 | 0.33 |
ENSDART00000185618
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr1_-_55166511 | 0.32 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr21_+_29179887 | 0.27 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr21_-_7995994 | 0.24 |
ENSDART00000151632
|
zgc:171965
|
zgc:171965 |
| chr18_+_1145571 | 0.23 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr11_-_438492 | 0.23 |
ENSDART00000137121
ENSDART00000133784 |
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr17_+_3390772 | 0.21 |
ENSDART00000160073
|
sntg2
|
syntrophin, gamma 2 |
| chr11_-_438294 | 0.10 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr5_+_23136544 | 0.07 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr3_-_19200571 | 0.06 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr17_+_26803470 | 0.05 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr1_+_41690402 | 0.04 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr4_-_17278904 | 0.01 |
ENSDART00000178686
ENSDART00000135730 |
lrmp
|
lymphoid-restricted membrane protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of cxxc1a+cxxc1b+mbd1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |