Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
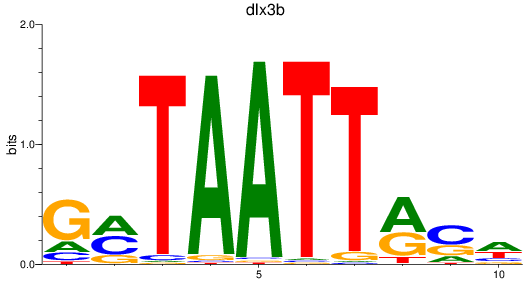
Results for dlx3b
Z-value: 0.39
Transcription factors associated with dlx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx3b
|
ENSDARG00000014626 | distal-less homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx3b | dr11_v1_chr12_+_5708400_5708400 | 0.04 | 7.1e-01 | Click! |
Activity profile of dlx3b motif
Sorted Z-values of dlx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_17023392 | 1.72 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr16_+_5774977 | 1.67 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr14_-_36378494 | 1.46 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr24_+_22485710 | 1.42 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr18_-_1185772 | 1.31 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr24_-_17029374 | 1.27 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr6_-_35472923 | 1.20 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr3_-_21280373 | 1.17 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr18_+_41232719 | 1.14 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr16_-_12953739 | 1.06 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr7_+_30787903 | 1.03 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr10_-_34871737 | 0.97 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr1_-_58036509 | 0.95 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr21_+_6751760 | 0.95 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr16_-_19890303 | 0.90 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr4_-_17629444 | 0.90 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr21_+_6751405 | 0.89 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr19_-_32641725 | 0.80 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr14_-_34044369 | 0.78 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr20_-_27225876 | 0.78 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr14_-_30587814 | 0.76 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr7_-_25895189 | 0.76 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr4_-_15420452 | 0.73 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr15_-_755023 | 0.73 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
| chr23_+_26946744 | 0.71 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr21_-_5856050 | 0.70 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr14_-_24410673 | 0.69 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr23_-_20051369 | 0.66 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr24_-_29963858 | 0.65 |
ENSDART00000183442
|
CR352310.1
|
|
| chr22_-_7129631 | 0.64 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr10_+_22381802 | 0.64 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr18_-_2433011 | 0.62 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr9_-_32730487 | 0.61 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr17_+_44780166 | 0.61 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr23_+_36130883 | 0.61 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr16_+_42471455 | 0.60 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr21_-_43949208 | 0.58 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_+_11532025 | 0.58 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr11_+_38280454 | 0.57 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr21_-_10488379 | 0.57 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr19_+_20793237 | 0.56 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr23_+_28582865 | 0.54 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr21_-_26490186 | 0.54 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr14_+_36220479 | 0.54 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr9_-_21976670 | 0.53 |
ENSDART00000104322
|
uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr20_+_30490682 | 0.52 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr17_-_15546862 | 0.52 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr2_-_55298075 | 0.51 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr6_-_6487876 | 0.51 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr25_-_32869794 | 0.50 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr7_-_54679595 | 0.49 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr15_-_9272328 | 0.48 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr5_+_27525477 | 0.48 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr1_+_37752171 | 0.48 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr11_-_6974022 | 0.48 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr22_-_3564563 | 0.47 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr23_-_7799184 | 0.45 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr10_+_21867307 | 0.42 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr19_+_20793388 | 0.41 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr15_-_42736433 | 0.41 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr2_+_2168547 | 0.41 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr11_+_18873113 | 0.41 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr4_-_9891874 | 0.40 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr11_-_34522249 | 0.40 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr2_-_23004286 | 0.39 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr16_-_13623928 | 0.38 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr2_-_32826108 | 0.37 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr7_+_30926289 | 0.37 |
ENSDART00000173518
|
si:dkey-1h4.4
|
si:dkey-1h4.4 |
| chr7_-_69647988 | 0.37 |
ENSDART00000169943
|
LO018231.1
|
|
| chr11_+_18873619 | 0.37 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr4_+_14727018 | 0.36 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr3_-_28428198 | 0.35 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr20_-_9462433 | 0.35 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr11_-_19775182 | 0.35 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr24_-_25004553 | 0.35 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr12_+_22580579 | 0.35 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr1_-_44701313 | 0.35 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr16_-_13623659 | 0.34 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr22_-_24284447 | 0.34 |
ENSDART00000149894
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr14_+_23717165 | 0.34 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_-_42607451 | 0.33 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr1_+_10318089 | 0.33 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr18_-_19095141 | 0.32 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr16_-_29194517 | 0.32 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr19_+_22062202 | 0.32 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr2_+_35421285 | 0.32 |
ENSDART00000143787
|
tnr
|
tenascin R (restrictin, janusin) |
| chr21_+_15824182 | 0.31 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr8_+_28452738 | 0.31 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr8_-_31107537 | 0.31 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr11_-_39105253 | 0.31 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr14_+_25817628 | 0.31 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr16_-_21620947 | 0.29 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_17346502 | 0.28 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_+_32832538 | 0.28 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr11_+_30244356 | 0.27 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr15_+_5360407 | 0.27 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr5_+_63857055 | 0.27 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr6_+_9289802 | 0.26 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_-_28671139 | 0.26 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr17_-_20717845 | 0.25 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr7_+_15872357 | 0.24 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr18_-_46258612 | 0.21 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr15_-_18115540 | 0.21 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr1_+_23783349 | 0.20 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr4_+_14727212 | 0.20 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr16_-_13613475 | 0.20 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr9_+_11034314 | 0.19 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr23_-_16682186 | 0.19 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr1_-_47071979 | 0.19 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chrM_+_9052 | 0.18 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr18_-_37007294 | 0.18 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr21_+_30355767 | 0.18 |
ENSDART00000189948
|
CR749164.1
|
|
| chr25_+_6471401 | 0.18 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr4_+_3980247 | 0.17 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_+_18588551 | 0.17 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr11_-_42554290 | 0.16 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr8_-_5847533 | 0.16 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr23_-_18130264 | 0.16 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr10_+_32104305 | 0.16 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr17_+_49500820 | 0.16 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr20_-_35578435 | 0.16 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr3_-_46811611 | 0.15 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_50461981 | 0.15 |
ENSDART00000158761
|
CU896640.1
|
|
| chr16_-_16120941 | 0.15 |
ENSDART00000131227
|
ankib1b
|
ankyrin repeat and IBR domain containing 1b |
| chr21_-_37790727 | 0.15 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr4_+_57580303 | 0.14 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr20_+_13783040 | 0.14 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr4_+_22343093 | 0.14 |
ENSDART00000023588
|
guca1a
|
guanylate cyclase activator 1A |
| chr5_-_51619742 | 0.14 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr8_-_23776399 | 0.13 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr12_-_19007834 | 0.13 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr12_+_25432627 | 0.13 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr19_+_31904836 | 0.12 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr13_+_22295905 | 0.12 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr4_+_41311345 | 0.12 |
ENSDART00000151905
|
si:dkey-16p19.8
|
si:dkey-16p19.8 |
| chr5_+_56268436 | 0.11 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr19_-_32914227 | 0.11 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr4_+_12612723 | 0.11 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr7_-_12464412 | 0.11 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr19_-_38830582 | 0.10 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr12_-_35386910 | 0.10 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr4_+_49322310 | 0.10 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr8_-_24252933 | 0.10 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr3_+_22059066 | 0.09 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr2_+_56213694 | 0.09 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr18_-_37007061 | 0.09 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr25_+_10410620 | 0.09 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr20_-_37813863 | 0.09 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr15_-_33172246 | 0.09 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr11_-_42418374 | 0.08 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr3_-_31079186 | 0.08 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr2_-_26720854 | 0.08 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr8_-_15129573 | 0.08 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr16_+_31804590 | 0.08 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr20_+_4060839 | 0.08 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr8_-_43997538 | 0.07 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr14_-_32631013 | 0.07 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr2_+_35612621 | 0.07 |
ENSDART00000143082
|
si:dkey-4i23.5
|
si:dkey-4i23.5 |
| chr25_-_18913336 | 0.07 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr8_-_39822917 | 0.07 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr14_-_7207961 | 0.07 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_-_20312146 | 0.06 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr6_-_40713183 | 0.06 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr16_-_21038015 | 0.05 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr11_+_13224281 | 0.05 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr11_-_40147032 | 0.03 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr12_-_6818676 | 0.03 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr20_+_23440632 | 0.02 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr6_+_60004871 | 0.02 |
ENSDART00000179832
|
CABZ01079267.1
|
|
| chr23_-_19230627 | 0.02 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr13_+_35339182 | 0.02 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr22_+_21324398 | 0.02 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr1_+_57311901 | 0.01 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr14_-_33821515 | 0.01 |
ENSDART00000173218
|
vimr2
|
vimentin-related 2 |
| chr2_+_32826235 | 0.01 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr11_+_33818179 | 0.00 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr11_+_33312601 | 0.00 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr10_-_21955231 | 0.00 |
ENSDART00000183695
|
FO744833.2
|
|
| chr20_-_32270866 | 0.00 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr5_-_27438812 | 0.00 |
ENSDART00000078755
|
drd7
|
dopamine receptor D7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 1.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 1.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.6 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.6 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 1.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.4 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.7 | GO:0003146 | heart jogging(GO:0003146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 1.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.0 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |