Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
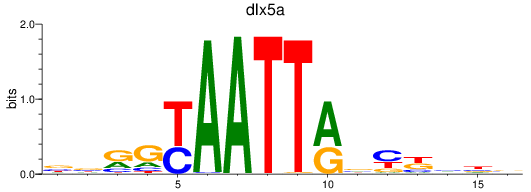
Results for dlx5a
Z-value: 0.64
Transcription factors associated with dlx5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx5a
|
ENSDARG00000042296 | distal-less homeobox 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx5a | dr11_v1_chr19_-_41472228_41472228 | -0.46 | 3.5e-06 | Click! |
Activity profile of dlx5a motif
Sorted Z-values of dlx5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_29955368 | 7.39 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr17_+_43867889 | 6.05 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr17_+_43863708 | 5.91 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr10_+_19596214 | 4.61 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr7_+_69019851 | 4.02 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr21_+_6751405 | 3.69 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr21_+_6751760 | 3.69 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr2_+_6253246 | 3.40 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr11_-_44801968 | 3.26 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr6_+_3680651 | 2.57 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr11_+_23760470 | 2.52 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr10_-_26512742 | 2.28 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr23_+_40460333 | 2.24 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr23_+_2906031 | 1.95 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr20_-_14114078 | 1.93 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr13_+_24829996 | 1.92 |
ENSDART00000147194
|
si:ch211-223p8.8
|
si:ch211-223p8.8 |
| chr16_+_35661331 | 1.89 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr10_-_40968095 | 1.86 |
ENSDART00000184104
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr10_-_26512993 | 1.85 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr13_-_33227411 | 1.74 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr16_-_42056137 | 1.67 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr16_+_35661771 | 1.65 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr21_+_39948300 | 1.63 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr23_+_33963619 | 1.51 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr17_+_41343167 | 1.43 |
ENSDART00000156405
|
mocs1
|
molybdenum cofactor synthesis 1 |
| chr8_-_50888806 | 1.42 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr16_+_45425181 | 1.36 |
ENSDART00000168591
|
phf1
|
PHD finger protein 1 |
| chr5_-_7829657 | 1.28 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr13_-_31435137 | 1.23 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr19_+_5480327 | 1.21 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr24_+_1023839 | 1.20 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr24_-_39637418 | 1.19 |
ENSDART00000142522
|
fam234a
|
family with sequence similarity 234, member A |
| chr6_+_23026714 | 1.17 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr7_-_40110140 | 1.14 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr24_+_7336411 | 1.12 |
ENSDART00000191170
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr6_-_6487876 | 1.04 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr15_+_15856178 | 0.98 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr18_+_11970987 | 0.94 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr20_-_47188966 | 0.91 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr17_+_25187226 | 0.90 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr15_-_44512461 | 0.88 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr20_-_2949028 | 0.83 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr3_-_46817499 | 0.82 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr2_-_21819421 | 0.78 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr2_-_33645411 | 0.76 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr9_-_49964810 | 0.76 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr2_-_2451181 | 0.75 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr4_-_5795309 | 0.75 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr16_+_45424907 | 0.75 |
ENSDART00000134471
|
phf1
|
PHD finger protein 1 |
| chr2_+_38370535 | 0.74 |
ENSDART00000179614
|
psmb11a
|
proteasome subunit beta 11a |
| chr20_+_39457598 | 0.74 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr7_-_12464412 | 0.69 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr11_+_30057762 | 0.68 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr23_-_31913231 | 0.67 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_-_25895189 | 0.65 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr15_+_5360407 | 0.61 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr16_+_42471455 | 0.56 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr5_-_30620625 | 0.53 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr1_-_53277413 | 0.51 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr18_+_50461981 | 0.50 |
ENSDART00000158761
|
CU896640.1
|
|
| chr20_+_14114258 | 0.47 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr11_+_31864921 | 0.46 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr24_-_31843173 | 0.45 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr16_+_54263921 | 0.45 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr10_+_2529037 | 0.44 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr23_+_34990693 | 0.44 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr3_+_17537352 | 0.43 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr17_+_49500820 | 0.41 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr1_-_17715493 | 0.38 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr2_-_10896745 | 0.37 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr16_+_31804590 | 0.37 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr14_+_11290828 | 0.34 |
ENSDART00000184078
|
rlim
|
ring finger protein, LIM domain interacting |
| chr4_+_306036 | 0.33 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr23_+_31912882 | 0.31 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr13_+_255067 | 0.29 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr23_-_31913069 | 0.28 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr25_-_34632050 | 0.25 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr3_-_23512285 | 0.24 |
ENSDART00000159151
|
BX682558.1
|
|
| chr2_-_17694504 | 0.23 |
ENSDART00000133709
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr17_-_43863700 | 0.21 |
ENSDART00000157530
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr4_-_32466676 | 0.20 |
ENSDART00000193347
ENSDART00000169396 ENSDART00000145784 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr18_+_28106139 | 0.17 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr6_-_40922971 | 0.17 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr1_+_40199074 | 0.16 |
ENSDART00000179679
ENSDART00000136849 |
si:ch211-113e8.5
|
si:ch211-113e8.5 |
| chr19_+_4800549 | 0.16 |
ENSDART00000056235
|
st3gal1l
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like |
| chr4_-_71153608 | 0.14 |
ENSDART00000190144
ENSDART00000158406 |
si:dkey-193i10.4
|
si:dkey-193i10.4 |
| chr25_-_7705487 | 0.12 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr8_-_39822917 | 0.11 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr19_+_19512515 | 0.11 |
ENSDART00000180065
|
jazf1a
|
JAZF zinc finger 1a |
| chr3_+_32832538 | 0.11 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr25_-_26893006 | 0.10 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr15_+_1796313 | 0.09 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr5_+_66433287 | 0.06 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr25_-_18913336 | 0.02 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr10_-_13343831 | 0.02 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_43891051 | 0.01 |
ENSDART00000024213
|
cxcr5
|
chemokine (C-X-C motif) receptor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 5.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 3.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 2.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.1 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.1 | 0.5 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.3 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 7.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 1.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.8 | GO:0048796 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 1.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 2.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.9 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 1.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 8.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 5.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.9 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 9.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 1.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 3.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide hormone activity(GO:0005184) neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |