Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
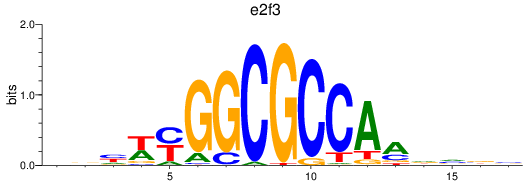
Results for e2f3
Z-value: 0.40
Transcription factors associated with e2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f3
|
ENSDARG00000070463 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f3 | dr11_v1_chr19_-_29294457_29294457 | -0.30 | 3.4e-03 | Click! |
Activity profile of e2f3 motif
Sorted Z-values of e2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_186583 | 3.35 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr24_+_39108243 | 3.15 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr6_+_60112200 | 2.18 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr18_+_54354 | 2.17 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr9_-_2892045 | 1.88 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr3_-_53486169 | 1.80 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr9_-_2892250 | 1.79 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr15_+_33991928 | 1.68 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr17_-_29224908 | 1.65 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr14_+_22076596 | 1.53 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr24_+_21684189 | 1.50 |
ENSDART00000014696
|
pdx1
|
pancreatic and duodenal homeobox 1 |
| chr15_-_34056733 | 1.39 |
ENSDART00000170130
ENSDART00000188272 |
VWDE
|
si:dkey-30e9.7 |
| chr19_+_636886 | 1.39 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr19_+_43604643 | 1.38 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr16_-_25085327 | 1.12 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr23_+_43950674 | 1.07 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr14_+_904850 | 1.03 |
ENSDART00000161847
|
si:ch73-208h1.1
|
si:ch73-208h1.1 |
| chr16_-_21140097 | 1.02 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr18_+_34861568 | 1.01 |
ENSDART00000192825
|
LO018333.1
|
|
| chr20_-_44090624 | 1.00 |
ENSDART00000048978
ENSDART00000082283 ENSDART00000082276 |
runx2b
|
runt-related transcription factor 2b |
| chr19_+_43604256 | 0.99 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr11_-_11792766 | 0.95 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr17_-_6535941 | 0.91 |
ENSDART00000109249
|
cenpo
|
centromere protein O |
| chr19_+_2546775 | 0.90 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr7_+_26534131 | 0.88 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr19_-_6988837 | 0.85 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr21_-_26490186 | 0.80 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr23_-_18913032 | 0.71 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr22_-_20259309 | 0.66 |
ENSDART00000139160
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr7_+_67434677 | 0.65 |
ENSDART00000165971
ENSDART00000166702 |
kars
|
lysyl-tRNA synthetase |
| chr25_+_36339867 | 0.64 |
ENSDART00000152195
|
si:ch211-113a14.18
|
si:ch211-113a14.18 |
| chr1_-_42289704 | 0.64 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr15_-_37425468 | 0.61 |
ENSDART00000059630
|
si:ch211-113j13.2
|
si:ch211-113j13.2 |
| chr15_-_14884332 | 0.52 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr11_-_44945636 | 0.50 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr4_-_72100774 | 0.49 |
ENSDART00000170099
|
slco1f1
|
solute carrier organic anion transporter family, member 1F1 |
| chr16_+_28728347 | 0.48 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr6_-_1865323 | 0.47 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr14_-_30918662 | 0.44 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr21_-_21514176 | 0.42 |
ENSDART00000031205
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr15_-_2954443 | 0.38 |
ENSDART00000161053
|
zgc:153184
|
zgc:153184 |
| chr23_-_44219902 | 0.32 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr4_-_9909371 | 0.31 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr3_-_5664123 | 0.28 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr7_-_1101071 | 0.20 |
ENSDART00000176053
|
dctn1a
|
dynactin 1a |
| chr5_+_66433287 | 0.15 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr25_+_6451038 | 0.14 |
ENSDART00000009971
|
snx33
|
sorting nexin 33 |
| chr6_+_7533601 | 0.14 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr14_+_14841685 | 0.11 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr17_-_465285 | 0.10 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr8_-_11988065 | 0.09 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr3_-_32275975 | 0.09 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr3_-_61592417 | 0.09 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr16_+_40575742 | 0.08 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr17_+_6536152 | 0.06 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr13_+_13033837 | 0.01 |
ENSDART00000079558
|
letm1
|
leucine zipper-EF-hand containing transmembrane protein 1 |
| chr14_-_31814149 | 0.00 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_38540411 | 0.00 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 3.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 3.7 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 1.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 1.0 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.7 | GO:0021782 | glial cell development(GO:0021782) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.9 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 2.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 3.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.3 | 2.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |