Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
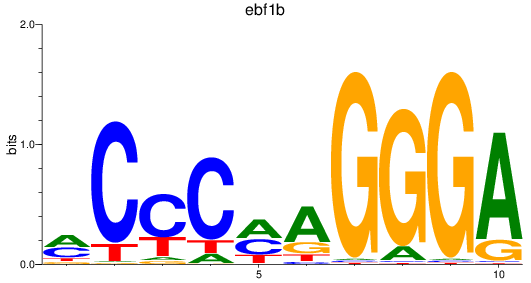
Results for ebf1b
Z-value: 1.07
Transcription factors associated with ebf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf1b
|
ENSDARG00000069196 | EBF transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf1b | dr11_v1_chr21_-_33851877_33851877 | 0.69 | 7.9e-15 | Click! |
Activity profile of ebf1b motif
Sorted Z-values of ebf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_34933183 | 19.12 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr12_-_35787801 | 13.52 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr4_+_11375894 | 12.76 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr21_-_19018455 | 12.30 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr10_+_19554604 | 10.78 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr25_+_21324588 | 10.44 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr14_+_25817246 | 10.33 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr19_-_13733870 | 9.88 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr3_+_49397115 | 9.87 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr9_+_44430974 | 8.75 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr4_+_17336557 | 8.34 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr5_-_37933983 | 8.31 |
ENSDART00000185364
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr10_+_21650828 | 8.18 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr7_+_58686860 | 8.03 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr7_-_52842007 | 7.97 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr21_+_26389391 | 7.89 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr1_-_44701313 | 6.83 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr10_+_33982010 | 6.60 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr23_+_26946744 | 6.58 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr13_-_40120252 | 6.27 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr9_+_44431174 | 6.23 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr4_+_74255904 | 5.97 |
ENSDART00000174385
|
ptprr
|
protein tyrosine phosphatase, receptor type, r |
| chr3_-_42086577 | 5.59 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr23_-_21471022 | 5.55 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_22831836 | 5.46 |
ENSDART00000142585
|
neb
|
nebulin |
| chr18_-_19095141 | 5.31 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr23_+_40460333 | 5.21 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr1_-_46989650 | 5.19 |
ENSDART00000129072
ENSDART00000150507 |
si:dkey-22n8.3
|
si:dkey-22n8.3 |
| chr13_+_17702853 | 4.91 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr14_+_25817628 | 4.88 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr1_+_41666611 | 4.86 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr12_-_33359654 | 4.78 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_-_2196267 | 4.78 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr12_-_26241810 | 4.76 |
ENSDART00000180335
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr18_+_50276653 | 4.74 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr4_+_13810811 | 4.70 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr4_-_5019113 | 4.52 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr16_-_25535430 | 4.45 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr23_+_20931030 | 4.40 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr15_+_6114109 | 4.40 |
ENSDART00000184937
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr4_+_20318127 | 4.33 |
ENSDART00000028856
ENSDART00000132909 |
cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr5_+_25952340 | 4.30 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr17_-_43558494 | 4.23 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr18_+_50276337 | 4.01 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr18_+_10820430 | 3.95 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr25_-_13105310 | 3.83 |
ENSDART00000172269
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr14_-_32405387 | 3.75 |
ENSDART00000184647
|
fgf13a
|
fibroblast growth factor 13a |
| chr21_-_2273244 | 3.69 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr13_-_32906265 | 3.67 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr23_-_5032587 | 3.66 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr1_+_41690402 | 3.61 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr13_+_17702522 | 3.56 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr15_-_30505607 | 3.54 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr24_+_1294176 | 3.49 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr4_+_37992753 | 3.45 |
ENSDART00000193890
|
CR759843.2
|
|
| chr16_-_47245563 | 3.39 |
ENSDART00000190225
|
LO017877.2
|
|
| chr2_+_38608290 | 3.30 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr11_+_31323746 | 3.13 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr24_-_25691020 | 3.11 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr13_+_1439152 | 2.88 |
ENSDART00000159047
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr19_-_26863626 | 2.85 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr11_+_31558006 | 2.79 |
ENSDART00000024296
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr4_-_10599062 | 2.78 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr10_-_31563049 | 2.77 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr6_+_58406014 | 2.71 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr5_+_57726425 | 2.64 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr10_+_26118122 | 2.63 |
ENSDART00000079207
|
trim47
|
tripartite motif containing 47 |
| chr3_+_62317990 | 2.63 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr24_-_12749116 | 2.62 |
ENSDART00000093583
|
cox19
|
COX19 cytochrome c oxidase assembly factor |
| chr18_+_33580391 | 2.61 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr3_+_36160979 | 2.51 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr15_+_5321612 | 2.50 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr5_+_457729 | 2.47 |
ENSDART00000025183
|
ift74
|
intraflagellar transport 74 |
| chr9_+_36356740 | 2.46 |
ENSDART00000139033
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr21_-_14775724 | 2.44 |
ENSDART00000043239
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr14_-_33083539 | 2.38 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr24_+_23730061 | 2.36 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr13_+_1928361 | 2.35 |
ENSDART00000164764
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr1_+_49878000 | 2.32 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr11_-_33857911 | 2.30 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr22_+_39058269 | 2.30 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr6_-_42036685 | 2.15 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr8_+_30671060 | 2.10 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr6_-_13709591 | 2.09 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr11_-_8782871 | 2.08 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr23_+_2825940 | 2.06 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr7_+_25920792 | 2.05 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr23_+_26946429 | 2.01 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr10_+_45059702 | 2.00 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr20_+_48782068 | 1.95 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr15_+_5339056 | 1.93 |
ENSDART00000174319
|
CABZ01018874.2
|
|
| chr14_-_41369629 | 1.93 |
ENSDART00000173040
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr9_+_21268739 | 1.90 |
ENSDART00000186514
|
BX511129.3
|
|
| chr22_+_2660505 | 1.90 |
ENSDART00000135555
ENSDART00000031485 |
znf1162
|
zinc finger protein 1162 |
| chr1_-_16394814 | 1.86 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr18_-_13360106 | 1.86 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr7_-_8961941 | 1.86 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr24_-_25184553 | 1.85 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr19_+_40335271 | 1.84 |
ENSDART00000135648
|
fam133b
|
family with sequence similarity 133, member B |
| chr18_+_20871142 | 1.71 |
ENSDART00000188274
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr2_+_5371492 | 1.69 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr17_+_6467398 | 1.68 |
ENSDART00000149527
|
COQ8A (1 of many)
|
si:dkey-36g24.3 |
| chr16_-_33076764 | 1.66 |
ENSDART00000192917
|
dopey1
|
dopey family member 1 |
| chr4_+_6840223 | 1.65 |
ENSDART00000161329
|
dock4b
|
dedicator of cytokinesis 4b |
| chr1_+_13930625 | 1.60 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr21_-_7687544 | 1.57 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr23_-_31506854 | 1.52 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr20_-_46467280 | 1.39 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr6_+_15145123 | 1.32 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr9_-_45149695 | 1.32 |
ENSDART00000162033
|
CU467837.1
|
|
| chr5_+_32141790 | 1.32 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chrM_+_3803 | 1.31 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr11_+_31558207 | 1.22 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr14_-_26094172 | 1.17 |
ENSDART00000150131
|
mtnr1al
|
melatonin receptor type 1A like |
| chr16_+_12281032 | 1.17 |
ENSDART00000138638
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr20_+_30378803 | 1.15 |
ENSDART00000148242
ENSDART00000169140 ENSDART00000062441 |
rnaseh1
|
ribonuclease H1 |
| chr10_-_27196093 | 1.15 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr12_-_11306766 | 1.14 |
ENSDART00000036989
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr7_-_41693004 | 1.14 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr10_+_10972795 | 1.14 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr8_-_52557455 | 1.13 |
ENSDART00000163596
|
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr12_+_23812530 | 1.11 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr16_-_30564319 | 1.06 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr13_-_39159810 | 1.03 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr7_-_32599669 | 1.02 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr15_-_23420079 | 0.94 |
ENSDART00000104525
ENSDART00000148840 |
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr18_-_4957022 | 0.91 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr12_-_36070473 | 0.85 |
ENSDART00000165720
|
gpr142
|
G protein-coupled receptor 142 |
| chr22_+_28446365 | 0.85 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr15_-_6976851 | 0.85 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr23_+_2760573 | 0.84 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr2_-_43123949 | 0.84 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr15_-_39819877 | 0.81 |
ENSDART00000142719
ENSDART00000139465 |
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr11_+_22134540 | 0.81 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr7_+_61876641 | 0.80 |
ENSDART00000165409
|
CR382383.2
|
|
| chr16_-_34212912 | 0.79 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr10_-_8041948 | 0.77 |
ENSDART00000059017
|
nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr22_+_567733 | 0.74 |
ENSDART00000171036
|
usp49
|
ubiquitin specific peptidase 49 |
| chr22_-_30881738 | 0.73 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr22_+_26290209 | 0.73 |
ENSDART00000060898
|
mrps28
|
mitochondrial ribosomal protein S28 |
| chr8_+_22916536 | 0.71 |
ENSDART00000136403
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr11_+_25638172 | 0.71 |
ENSDART00000114226
ENSDART00000143677 |
grm6b
|
glutamate receptor, metabotropic 6b |
| chr17_+_24597001 | 0.70 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr15_-_23936276 | 0.67 |
ENSDART00000137569
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr9_+_22080122 | 0.65 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr3_+_32691275 | 0.63 |
ENSDART00000191838
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr11_+_8660158 | 0.62 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr25_+_35020529 | 0.62 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr17_+_24222190 | 0.62 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr12_-_11560794 | 0.61 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr18_-_7112882 | 0.60 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr1_+_5275811 | 0.59 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr6_-_30863046 | 0.55 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr16_-_43139251 | 0.53 |
ENSDART00000021145
ENSDART00000165293 |
slc25a40
|
solute carrier family 25, member 40 |
| chr3_-_51142942 | 0.53 |
ENSDART00000160789
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr23_-_6920107 | 0.50 |
ENSDART00000149230
ENSDART00000182946 |
si:ch211-117c9.2
|
si:ch211-117c9.2 |
| chr2_-_30659222 | 0.48 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr3_-_46403778 | 0.48 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr12_+_13652747 | 0.48 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr22_-_7809904 | 0.47 |
ENSDART00000182269
ENSDART00000097201 ENSDART00000138522 |
si:ch73-44m9.3
si:ch73-44m9.1
|
si:ch73-44m9.3 si:ch73-44m9.1 |
| chr8_-_14067517 | 0.46 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr18_-_210478 | 0.46 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr23_+_45027263 | 0.45 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr6_-_6254432 | 0.41 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr2_-_32457919 | 0.39 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr1_-_50438247 | 0.33 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr16_-_48914757 | 0.33 |
ENSDART00000166740
|
FQ976914.1
|
|
| chr15_-_23482088 | 0.31 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr2_-_37896646 | 0.31 |
ENSDART00000075931
|
hbl1
|
hexose-binding lectin 1 |
| chr8_+_12930216 | 0.23 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr15_-_3466015 | 0.23 |
ENSDART00000169559
|
cog6
|
component of oligomeric golgi complex 6 |
| chr5_-_42083363 | 0.23 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr15_+_11427620 | 0.22 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr5_+_46481586 | 0.22 |
ENSDART00000144973
|
vcana
|
versican a |
| chr20_+_25711718 | 0.14 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr21_+_16144014 | 0.14 |
ENSDART00000163267
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr10_-_32639939 | 0.13 |
ENSDART00000126952
ENSDART00000099853 |
umodl1
|
uromodulin-like 1 |
| chr8_+_8196087 | 0.12 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr7_-_38806186 | 0.09 |
ENSDART00000173669
|
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr5_-_29112956 | 0.08 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr8_-_34762163 | 0.07 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr6_-_43922813 | 0.05 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr11_+_3575963 | 0.05 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr22_-_28373698 | 0.02 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr16_-_14534724 | 0.02 |
ENSDART00000148126
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 2.3 | 13.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 2.1 | 12.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 2.0 | 7.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.3 | 12.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.1 | 4.4 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.9 | 5.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.9 | 4.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.9 | 4.4 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.7 | 4.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.7 | 2.1 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.6 | 19.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.6 | 2.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 2.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 5.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 2.5 | GO:0035776 | intraciliary transport involved in cilium morphogenesis(GO:0035735) pronephric proximal tubule development(GO:0035776) |
| 0.3 | 2.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 4.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 8.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 4.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.3 | 3.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 2.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 6.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 3.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 3.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 2.8 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.2 | 3.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 7.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 1.9 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 4.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 5.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 10.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.9 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 1.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 1.5 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 4.3 | GO:0055017 | cardiac muscle tissue growth(GO:0055017) |
| 0.2 | 0.9 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.2 | 16.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 9.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 2.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 3.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.0 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 7.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.2 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) regulation of renal system process(GO:0098801) |
| 0.1 | 1.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 15.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 2.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 5.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 1.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.7 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 5.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 4.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 4.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 7.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 3.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 6.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 2.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 2.7 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 1.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.0 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 2.5 | 12.3 | GO:0005883 | neurofilament(GO:0005883) |
| 1.3 | 12.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.6 | 10.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 2.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 4.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 15.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 5.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 3.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 6.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 8.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 13.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 10.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 3.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 8.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 5.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 3.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 4.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 8.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 15.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.9 | 2.6 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.7 | 12.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.7 | 2.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.6 | 5.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 8.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.6 | 2.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.6 | 7.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 2.4 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.6 | 1.7 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.5 | 19.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 6.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 7.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 15.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 2.4 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 2.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 3.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 8.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 4.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 4.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 5.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 7.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 8.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 9.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 3.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 4.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 2.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 3.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 9.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 5.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 8.4 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 3.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 7.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 9.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 9.5 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 15.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 2.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 3.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 4.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 5.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 8.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |