Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
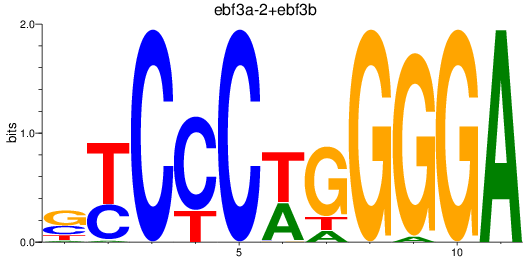
Results for ebf3a-2+ebf3b
Z-value: 0.92
Transcription factors associated with ebf3a-2+ebf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf3b
|
ENSDARG00000091287 | EBF transcription factor 3b |
|
ebf3a-2
|
ENSDARG00000100244 | EBF transcription factor 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf3a | dr11_v1_chr12_+_42436328_42436328 | 0.57 | 1.7e-09 | Click! |
| ebf3b | dr11_v1_chr17_-_20229812_20229812 | 0.29 | 5.1e-03 | Click! |
Activity profile of ebf3a-2+ebf3b motif
Sorted Z-values of ebf3a-2+ebf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_37559570 | 9.96 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr20_+_34933183 | 8.42 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr8_+_31821396 | 7.92 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr21_+_5129513 | 7.18 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr15_-_24869826 | 6.36 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr1_-_21409877 | 5.54 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr23_-_21471022 | 5.45 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr2_+_5371492 | 5.43 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr23_-_5683147 | 5.39 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr12_+_6195191 | 5.38 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr8_-_1051438 | 5.32 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr25_+_21324588 | 5.10 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr13_-_31452516 | 5.06 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr9_-_7673856 | 5.06 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr23_+_21455152 | 5.02 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr10_-_25769334 | 4.97 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr22_+_11756040 | 4.38 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr7_+_38811800 | 4.35 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr15_+_1372343 | 4.24 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr9_-_31278048 | 4.15 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr6_-_40697585 | 3.93 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr17_+_17955063 | 3.89 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr17_+_45454943 | 3.82 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr6_+_45932276 | 3.78 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr24_+_3972260 | 3.72 |
ENSDART00000131753
|
pfkpa
|
phosphofructokinase, platelet a |
| chr3_+_13559199 | 3.42 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr17_-_15611744 | 3.39 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr15_+_16521785 | 3.31 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr9_-_98982 | 3.27 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr19_-_8880688 | 3.22 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr16_-_32013913 | 3.07 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr3_-_28665291 | 2.99 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr3_+_50310684 | 2.98 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr17_+_45602640 | 2.96 |
ENSDART00000153822
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr7_+_66822229 | 2.95 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr9_-_36924388 | 2.92 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr3_+_16722014 | 2.74 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr25_-_24074500 | 2.73 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr15_-_1941042 | 2.69 |
ENSDART00000112946
ENSDART00000155504 |
dock10
|
dedicator of cytokinesis 10 |
| chr25_-_20523927 | 2.67 |
ENSDART00000114448
|
si:ch211-269c21.2
|
si:ch211-269c21.2 |
| chr25_+_35706493 | 2.66 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr16_-_9869056 | 2.62 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr2_-_30987538 | 2.61 |
ENSDART00000148157
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr20_-_53321499 | 2.58 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr17_+_38566717 | 2.46 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr13_-_13751017 | 2.46 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr5_-_25174420 | 2.44 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr4_-_9008846 | 2.43 |
ENSDART00000164632
ENSDART00000091876 |
scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr21_-_24632778 | 2.40 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr2_-_39675829 | 2.31 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr5_-_19052184 | 2.29 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr2_-_37465517 | 2.28 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr22_+_11775269 | 2.21 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr7_-_52558495 | 2.21 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr6_+_55277419 | 2.17 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr24_+_4977862 | 2.12 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr2_-_32551178 | 2.08 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr13_+_17702853 | 2.04 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr17_+_53425730 | 2.04 |
ENSDART00000127617
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr23_-_5032587 | 2.03 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr2_+_38608290 | 1.97 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr6_+_2951645 | 1.95 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr7_-_18554603 | 1.94 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr16_-_47483142 | 1.91 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr20_-_25669813 | 1.87 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr6_-_49673476 | 1.83 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr11_-_27501027 | 1.83 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr1_+_40562540 | 1.80 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr12_-_28548789 | 1.76 |
ENSDART00000153062
|
si:ch73-180n10.1
|
si:ch73-180n10.1 |
| chr10_-_42237304 | 1.75 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr6_+_7250824 | 1.73 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr24_-_10828560 | 1.71 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr13_-_31622195 | 1.70 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr17_+_24222190 | 1.67 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr8_+_30671060 | 1.66 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr16_-_36093776 | 1.65 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr1_+_49878000 | 1.61 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr25_+_34915576 | 1.60 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr8_+_29856013 | 1.60 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr11_-_31039533 | 1.58 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr17_+_25955003 | 1.58 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr16_+_20198064 | 1.57 |
ENSDART00000006037
|
colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr9_-_33081781 | 1.55 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr16_-_35952789 | 1.53 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr4_+_37992753 | 1.51 |
ENSDART00000193890
|
CR759843.2
|
|
| chr18_-_47103131 | 1.51 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr24_+_4978055 | 1.47 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr11_+_13071645 | 1.47 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr21_-_38853737 | 1.47 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr6_+_45692026 | 1.44 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr23_-_16692312 | 1.43 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr23_-_36003282 | 1.42 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr2_-_55317035 | 1.39 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr17_+_28533102 | 1.36 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr16_-_34212912 | 1.36 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr11_-_25324534 | 1.34 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr21_+_21374277 | 1.30 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr13_+_17702522 | 1.27 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr2_-_56649883 | 1.26 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr4_+_5193198 | 1.22 |
ENSDART00000067388
|
fgf23
|
fibroblast growth factor 23 |
| chr11_+_7580079 | 1.21 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr19_+_9459050 | 1.19 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr4_-_28958601 | 1.19 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr17_-_45247151 | 1.18 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr10_-_5022176 | 1.17 |
ENSDART00000148277
|
hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr25_-_21716326 | 1.16 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr24_-_13364311 | 1.15 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr20_+_18209895 | 1.13 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr24_+_32472155 | 1.13 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr14_-_2196267 | 1.12 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr7_-_33949246 | 1.11 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr7_+_39410180 | 1.06 |
ENSDART00000168641
|
CT030188.1
|
|
| chr7_-_52334840 | 1.06 |
ENSDART00000174173
|
CR938716.1
|
|
| chr5_+_57726425 | 1.03 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr10_-_5016997 | 1.02 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr5_+_27267186 | 1.01 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr4_-_30712588 | 1.00 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr21_-_37194365 | 0.98 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr16_-_47245563 | 0.97 |
ENSDART00000190225
|
LO017877.2
|
|
| chr17_-_21441464 | 0.95 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr5_+_32141790 | 0.93 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr7_-_40630698 | 0.91 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr22_-_10470663 | 0.91 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr9_+_23714406 | 0.89 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr3_-_55649319 | 0.87 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr2_-_10943093 | 0.87 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr24_+_1294176 | 0.87 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr6_-_42036685 | 0.87 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr22_-_23253252 | 0.87 |
ENSDART00000175556
|
lhx9
|
LIM homeobox 9 |
| chr1_+_16548733 | 0.85 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr2_-_32457919 | 0.78 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr23_+_21459263 | 0.78 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr18_+_9382847 | 0.78 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr7_+_39410393 | 0.77 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr17_+_13763425 | 0.75 |
ENSDART00000105175
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr12_-_46176115 | 0.75 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr8_-_25422186 | 0.74 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr19_-_336777 | 0.68 |
ENSDART00000138837
|
golph3l
|
golgi phosphoprotein 3-like |
| chr23_+_20931030 | 0.67 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr9_-_37637179 | 0.67 |
ENSDART00000188143
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr6_+_102506 | 0.67 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr22_+_2844865 | 0.66 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr6_+_58406014 | 0.64 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr25_+_34915762 | 0.61 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr6_+_51779863 | 0.60 |
ENSDART00000108534
|
tmem74b
|
transmembrane protein 74B |
| chr10_+_10972795 | 0.60 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr11_-_33857911 | 0.56 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr4_-_760560 | 0.53 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr10_+_11286741 | 0.52 |
ENSDART00000187822
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr19_+_1370504 | 0.50 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr3_-_25813426 | 0.50 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr4_-_9579299 | 0.47 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr14_+_22129096 | 0.46 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr24_-_27400017 | 0.45 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr5_+_23634665 | 0.44 |
ENSDART00000125161
|
cx39.9
|
connexin 39.9 |
| chr25_-_13549577 | 0.40 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr9_-_33063083 | 0.40 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr3_+_23247325 | 0.39 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr12_+_18906407 | 0.39 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr19_-_10043142 | 0.38 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr2_-_144393 | 0.37 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr11_+_6159595 | 0.37 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr21_-_37194669 | 0.34 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr4_-_4756349 | 0.33 |
ENSDART00000057507
|
ssbp1
|
single-stranded DNA binding protein 1 |
| chr13_+_1928361 | 0.31 |
ENSDART00000164764
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr18_-_210478 | 0.29 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr23_+_16948049 | 0.26 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr1_-_8963484 | 0.26 |
ENSDART00000188035
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr8_-_21988833 | 0.26 |
ENSDART00000167708
|
nphp4
|
nephronophthisis 4 |
| chr10_+_11286925 | 0.25 |
ENSDART00000143615
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr13_-_25408387 | 0.25 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr4_+_8638622 | 0.22 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr23_+_35672542 | 0.22 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr5_+_38900249 | 0.21 |
ENSDART00000097856
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr24_+_25794318 | 0.21 |
ENSDART00000143395
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr18_-_26781616 | 0.20 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr20_+_2039518 | 0.20 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr8_-_15182232 | 0.18 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr21_-_14775724 | 0.14 |
ENSDART00000043239
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr10_-_27199135 | 0.12 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr18_+_30441740 | 0.11 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr3_-_43646733 | 0.10 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr1_-_191971 | 0.10 |
ENSDART00000164091
ENSDART00000159237 |
grtp1a
|
growth hormone regulated TBC protein 1a |
| chr13_+_3954715 | 0.09 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr4_-_69127091 | 0.07 |
ENSDART00000136092
|
si:ch211-209j12.3
|
si:ch211-209j12.3 |
| chr23_+_19814371 | 0.06 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr13_-_17723417 | 0.04 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr19_+_19756425 | 0.03 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr15_+_35718354 | 0.03 |
ENSDART00000110003
|
nyap2a
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2a |
| chr17_-_22324727 | 0.03 |
ENSDART00000160341
|
CU104709.1
|
|
| chr20_-_33966148 | 0.02 |
ENSDART00000148111
|
selp
|
selectin P |
| chr8_+_21297217 | 0.01 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3a-2+ebf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.2 | 7.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.9 | 2.7 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.7 | 4.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.7 | 3.3 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.6 | 3.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.6 | 4.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.5 | 12.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.5 | 5.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 1.4 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.4 | 1.8 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.4 | 1.7 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.4 | 3.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 5.5 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.3 | 3.7 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 0.9 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.3 | 0.6 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.3 | 8.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.3 | 1.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 2.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 2.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 3.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 3.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 3.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.2 | GO:0030320 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 1.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 3.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.5 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.7 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 3.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 4.6 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 2.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 4.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.9 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 3.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.5 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 6.7 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 3.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.0 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 7.1 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.2 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 3.3 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 2.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 3.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 5.3 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 2.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 3.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 7.2 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 5.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 5.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 9.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 5.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 6.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 3.5 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 2.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 3.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.9 | 5.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.7 | 2.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.5 | 5.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 5.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 1.0 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.3 | 3.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 2.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 5.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 8.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 7.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 14.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 2.4 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 2.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 4.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 6.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 1.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |