Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
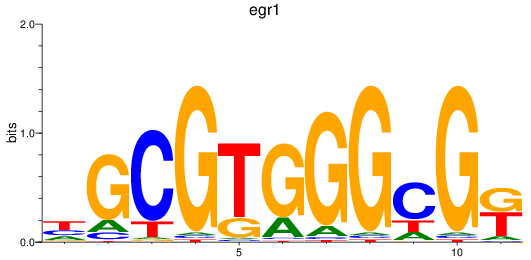
Results for egr1
Z-value: 1.01
Transcription factors associated with egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr1
|
ENSDARG00000037421 | early growth response 1 |
|
egr1
|
ENSDARG00000115563 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr1 | dr11_v1_chr14_-_21336385_21336385 | -0.25 | 1.4e-02 | Click! |
Activity profile of egr1 motif
Sorted Z-values of egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20422661 | 14.31 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr3_-_61185746 | 13.15 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr18_+_402048 | 11.53 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr25_-_37501371 | 10.44 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr17_+_53311618 | 10.22 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr18_+_5490668 | 8.95 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr13_+_9432501 | 8.40 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr7_+_74150839 | 8.17 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr11_+_77526 | 7.63 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr15_-_20939579 | 7.53 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr17_+_53311243 | 7.18 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr2_-_48753873 | 7.18 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr14_+_2243 | 6.95 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr3_-_5228137 | 6.77 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr10_+_252425 | 6.71 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr20_-_54381034 | 6.55 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr20_+_54356272 | 6.54 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr18_-_50845804 | 6.52 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr12_-_49149800 | 6.09 |
ENSDART00000182290
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr9_-_105135 | 6.07 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr24_-_42072886 | 6.05 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr11_-_44347587 | 5.95 |
ENSDART00000172919
|
si:cabz01100188.1
|
si:cabz01100188.1 |
| chr19_+_48102560 | 5.76 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr6_-_13206255 | 5.49 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr20_-_54377933 | 5.34 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr4_+_77948517 | 5.07 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_6193448 | 4.99 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr23_+_39606108 | 4.96 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr22_-_193234 | 4.91 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr10_-_34002185 | 4.90 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr18_-_49078428 | 4.85 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr15_+_3284684 | 4.79 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr12_-_49168398 | 4.64 |
ENSDART00000186608
|
FO704624.1
|
|
| chr15_-_47848544 | 4.54 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr15_-_17099560 | 4.54 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr3_+_16722014 | 4.53 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr6_+_14301214 | 4.52 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr21_-_233282 | 4.50 |
ENSDART00000157684
|
bxdc2
|
brix domain containing 2 |
| chr20_+_39250673 | 4.24 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr21_-_308852 | 4.16 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr12_+_48851381 | 4.03 |
ENSDART00000187241
ENSDART00000187796 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr24_+_42127983 | 3.85 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_+_35880600 | 3.83 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr14_+_164556 | 3.83 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr21_+_19368720 | 3.83 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_1211242 | 3.76 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr1_+_176583 | 3.75 |
ENSDART00000168760
ENSDART00000160425 |
LAMP1
|
lysosomal associated membrane protein 1 |
| chr17_-_2584423 | 3.70 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr5_+_72377851 | 3.68 |
ENSDART00000160479
|
lman2la
|
lectin, mannose-binding 2-like a |
| chr6_+_2093367 | 3.65 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr21_+_26620867 | 3.64 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr18_-_50956280 | 3.38 |
ENSDART00000058457
|
st7
|
suppression of tumorigenicity 7 |
| chr8_+_45294767 | 3.33 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr21_+_30950097 | 3.27 |
ENSDART00000187572
|
rhogb
|
ras homolog family member Gb |
| chr19_-_7225060 | 3.25 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr11_+_45461853 | 3.24 |
ENSDART00000173409
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr16_+_23978978 | 3.21 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr22_-_80533 | 3.15 |
ENSDART00000183124
|
CABZ01101813.1
|
|
| chr5_+_337215 | 2.97 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
| chr23_+_44374041 | 2.91 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr21_+_103194 | 2.86 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr8_-_53490376 | 2.81 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr8_-_14144707 | 2.78 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr20_+_7398090 | 2.71 |
ENSDART00000108649
|
pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr8_+_2642384 | 2.70 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr13_+_25486608 | 2.69 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr7_-_46019756 | 2.68 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr1_-_8653385 | 2.68 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr24_+_37406535 | 2.63 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr23_-_270847 | 2.62 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr25_-_32888115 | 2.56 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr20_-_48172556 | 2.53 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr7_+_46019780 | 2.50 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr25_-_16146851 | 2.47 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr11_+_1565806 | 2.46 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr11_+_45436703 | 2.34 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr9_-_23990416 | 2.33 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr2_+_59046036 | 2.33 |
ENSDART00000158860
|
stk11
|
serine/threonine kinase 11 |
| chr17_+_584369 | 2.32 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr11_+_2855430 | 2.32 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr14_+_12316581 | 2.25 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr6_+_13206516 | 2.24 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr3_-_34041878 | 2.20 |
ENSDART00000185325
ENSDART00000193934 |
ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr18_+_16246806 | 2.16 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr16_+_13965923 | 2.15 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr17_+_20639180 | 2.15 |
ENSDART00000063624
|
npy4r
|
neuropeptide Y receptor Y4 |
| chr5_+_37032038 | 2.14 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_-_54639964 | 2.14 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr4_+_30785713 | 2.13 |
ENSDART00000165945
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr14_+_7939398 | 2.13 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr12_-_20303438 | 2.12 |
ENSDART00000153057
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr8_+_20398445 | 2.08 |
ENSDART00000134755
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr20_+_38458084 | 2.06 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr13_-_50567658 | 2.06 |
ENSDART00000132281
|
blnk
|
B cell linker |
| chr23_+_45822935 | 2.02 |
ENSDART00000161892
|
vdra
|
vitamin D receptor a |
| chr17_+_27456804 | 2.01 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr10_+_8554929 | 1.94 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr10_-_35030673 | 1.93 |
ENSDART00000135338
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr1_+_56972616 | 1.89 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr24_+_19542323 | 1.87 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr15_+_3284416 | 1.87 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr3_+_19299309 | 1.86 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr16_+_19029297 | 1.86 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr1_-_59422880 | 1.82 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr2_+_105748 | 1.78 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr21_+_8198652 | 1.76 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr12_+_19030391 | 1.74 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr14_+_15155684 | 1.73 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr20_-_22778394 | 1.71 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr6_-_7776612 | 1.69 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr19_+_636886 | 1.69 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr11_+_45461419 | 1.67 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr1_-_44940830 | 1.67 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr20_-_26383368 | 1.65 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr23_-_27547931 | 1.64 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr1_-_49434798 | 1.61 |
ENSDART00000150842
ENSDART00000150789 |
ATP5MD
|
si:dkeyp-80c12.10 |
| chr6_-_43817761 | 1.61 |
ENSDART00000183945
|
foxp1b
|
forkhead box P1b |
| chr19_-_11237125 | 1.60 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr16_-_54405976 | 1.58 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr7_-_7904683 | 1.56 |
ENSDART00000192822
|
CABZ01030278.1
|
|
| chr4_+_63484571 | 1.55 |
ENSDART00000169518
ENSDART00000168681 |
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr16_-_13965518 | 1.54 |
ENSDART00000138304
|
si:dkey-85k15.4
|
si:dkey-85k15.4 |
| chr5_+_36513605 | 1.53 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr23_-_31060350 | 1.48 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr13_-_479129 | 1.48 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr19_-_703898 | 1.48 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr14_+_29769336 | 1.46 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr16_+_50163352 | 1.44 |
ENSDART00000153890
ENSDART00000060522 |
si:ch211-231m23.4
|
si:ch211-231m23.4 |
| chr17_+_53435279 | 1.43 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr10_-_40557210 | 1.42 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr20_-_51697437 | 1.40 |
ENSDART00000145391
|
si:ch211-14a11.2
|
si:ch211-14a11.2 |
| chr11_-_26666501 | 1.38 |
ENSDART00000188067
ENSDART00000111539 |
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr12_-_44016898 | 1.36 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr7_-_10560964 | 1.36 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr25_+_36315127 | 1.35 |
ENSDART00000191984
|
zgc:165555
|
zgc:165555 |
| chr20_-_52631998 | 1.35 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr25_-_36370292 | 1.35 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr15_-_14038227 | 1.33 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr19_+_2619444 | 1.27 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr20_-_46808870 | 1.27 |
ENSDART00000190911
|
esrrgb
|
estrogen-related receptor gamma b |
| chr10_-_40402904 | 1.27 |
ENSDART00000145101
|
taar20t
|
trace amine associated receptor 20t |
| chr15_+_47838836 | 1.25 |
ENSDART00000182918
|
CU682604.1
|
|
| chr5_+_66170479 | 1.25 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr9_+_45227028 | 1.19 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr20_-_10288156 | 1.19 |
ENSDART00000064110
|
si:dkey-63b1.1
|
si:dkey-63b1.1 |
| chr15_-_14486534 | 1.18 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_+_935565 | 1.15 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr16_-_26132122 | 1.15 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr1_-_48922273 | 1.11 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr11_+_37251825 | 1.11 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr8_+_28724692 | 1.11 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr25_+_37348730 | 1.10 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr1_+_58977262 | 1.08 |
ENSDART00000168832
|
eri2
|
ERI1 exoribonuclease family member 2 |
| chr10_+_40548616 | 1.08 |
ENSDART00000147894
|
si:ch211-238p8.10
|
si:ch211-238p8.10 |
| chr7_-_24995631 | 1.08 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr9_+_51784922 | 1.08 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr24_+_26379441 | 1.06 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr21_-_24865454 | 1.06 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr5_+_43870389 | 1.05 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr7_-_12596727 | 1.04 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr2_+_38271392 | 1.03 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr6_+_59808677 | 1.02 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr10_+_40774215 | 1.01 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr7_-_58776400 | 0.98 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr21_-_280769 | 0.97 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr14_+_17397534 | 0.96 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr10_-_40448736 | 0.91 |
ENSDART00000137644
ENSDART00000168190 |
taar20p
|
trace amine associated receptor 20p |
| chr25_+_1304173 | 0.91 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr9_-_41401564 | 0.91 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr17_+_34206167 | 0.89 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr7_-_66126628 | 0.87 |
ENSDART00000184492
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr10_+_2742499 | 0.87 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr16_+_52105227 | 0.86 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr23_-_22072460 | 0.85 |
ENSDART00000132343
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr22_-_20289948 | 0.84 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr19_+_43504480 | 0.83 |
ENSDART00000159421
|
BX649384.3
|
|
| chr2_+_50062165 | 0.83 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr24_-_30275735 | 0.82 |
ENSDART00000168723
|
snx7
|
sorting nexin 7 |
| chr5_-_1999417 | 0.80 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr22_+_24770744 | 0.79 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr19_-_24163845 | 0.77 |
ENSDART00000133277
|
prf1.2
|
perforin 1.2 |
| chr20_+_28803642 | 0.77 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr21_-_43398122 | 0.75 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr19_-_2115040 | 0.74 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr23_+_14269311 | 0.74 |
ENSDART00000170414
|
CR354537.1
|
|
| chr21_-_24865217 | 0.72 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr23_+_35426404 | 0.71 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr16_-_6821927 | 0.71 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr14_+_909769 | 0.69 |
ENSDART00000021346
ENSDART00000172777 |
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr1_-_411331 | 0.67 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr7_+_5968225 | 0.64 |
ENSDART00000083397
|
zgc:165555
|
zgc:165555 |
| chr19_-_3287393 | 0.63 |
ENSDART00000135698
|
si:ch211-133n4.10
|
si:ch211-133n4.10 |
| chr25_-_28587896 | 0.63 |
ENSDART00000111908
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr6_-_13498745 | 0.63 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr9_-_33121535 | 0.63 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr12_-_990149 | 0.63 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr4_+_70556298 | 0.62 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr8_-_1973220 | 0.60 |
ENSDART00000184435
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr16_-_12512568 | 0.58 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr20_+_32523576 | 0.58 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1902103 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 1.1 | 3.4 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 1.1 | 6.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.0 | 6.7 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.9 | 2.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.9 | 2.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.7 | 14.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.7 | 11.9 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.6 | 17.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.6 | 1.7 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.5 | 11.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.5 | 5.1 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.5 | 4.2 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.5 | 5.9 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.4 | 2.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.4 | 3.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 1.7 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.4 | 2.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.3 | 3.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 1.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.9 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 6.1 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) |
| 0.3 | 9.0 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.3 | 6.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 1.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 1.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 2.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 3.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.2 | 1.7 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 4.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 8.2 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.2 | 1.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 4.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 3.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 2.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.6 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.6 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 3.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.5 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.8 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 2.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 3.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 3.6 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 0.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 4.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 2.1 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) regulation of ERBB signaling pathway(GO:1901184) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.7 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 5.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 3.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 3.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 7.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.3 | GO:0010885 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.0 | 3.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 2.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.7 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.2 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 2.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.3 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 2.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.0 | GO:0007129 | synapsis(GO:0007129) protein sumoylation(GO:0016925) |
| 0.0 | 1.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 5.3 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 2.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 2.1 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 2.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 5.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.8 | 5.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 2.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.6 | 1.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.5 | 2.7 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 1.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.3 | 14.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 4.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 2.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.7 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 6.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 9.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 6.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 14.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 25.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 1.1 | 4.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.8 | 11.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 11.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.7 | 2.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 2.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 1.7 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.5 | 2.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.5 | 2.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 14.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 1.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 3.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 2.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 5.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 3.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 3.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 2.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.7 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.2 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 3.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 3.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 4.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 2.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 5.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 5.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 16.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 8.1 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 6.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 4.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 4.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 7.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 2.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 4.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 6.8 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 3.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 2.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 5.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 6.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 1.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 11.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.5 | 6.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 14.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 3.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 6.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 2.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 5.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 5.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 4.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 4.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.8 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 7.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |