Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
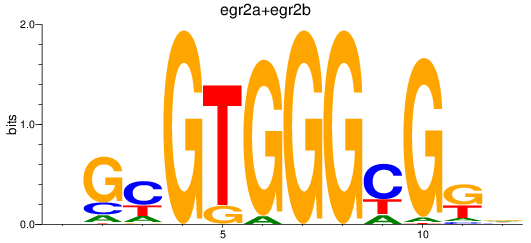
Results for egr2a+egr2b
Z-value: 1.12
Transcription factors associated with egr2a+egr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr2b
|
ENSDARG00000042826 | early growth response 2b |
|
egr2a
|
ENSDARG00000044098 | early growth response 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr2b | dr11_v1_chr12_-_8486330_8486330 | 0.54 | 2.3e-08 | Click! |
| egr2a | dr11_v1_chr17_-_43677471_43677471 | 0.19 | 6.9e-02 | Click! |
Activity profile of egr2a+egr2b motif
Sorted Z-values of egr2a+egr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20422661 | 13.53 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr2_-_22535 | 10.89 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr16_-_6821927 | 10.46 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr20_-_54462551 | 9.69 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr3_-_61185746 | 9.57 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr18_+_402048 | 7.76 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr5_-_52277643 | 7.04 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr2_+_33326522 | 6.93 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr10_-_32851847 | 6.56 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr21_+_5169154 | 6.52 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr11_+_1796426 | 6.48 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr6_-_15604157 | 6.48 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr11_+_7324704 | 6.36 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr21_+_28958471 | 6.30 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_+_38369872 | 6.28 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr6_-_15604417 | 6.23 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr11_-_2594045 | 6.01 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr24_+_2519761 | 5.95 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr2_+_47582681 | 5.92 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr19_+_342094 | 5.90 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr15_-_20916251 | 5.78 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_-_20303438 | 5.76 |
ENSDART00000153057
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr24_+_3963684 | 5.69 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr3_+_32492467 | 5.65 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr5_-_57655092 | 5.60 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr14_+_2243 | 5.55 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr17_-_20897250 | 5.50 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr21_-_43949208 | 5.49 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_-_52643970 | 5.46 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr25_-_207214 | 5.35 |
ENSDART00000193448
|
FP236318.3
|
|
| chr1_-_22861348 | 5.30 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr25_+_34984333 | 5.27 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr23_-_32156278 | 5.16 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_-_20939579 | 5.08 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr20_-_53366137 | 4.99 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr21_-_43952958 | 4.92 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr5_-_26093945 | 4.84 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr8_-_4618653 | 4.82 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr25_+_34013093 | 4.81 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr3_+_30921246 | 4.80 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr10_-_15919839 | 4.80 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr7_-_33023404 | 4.77 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr23_-_637347 | 4.75 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr23_-_39849155 | 4.70 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr15_+_1397811 | 4.50 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr25_-_12906872 | 4.49 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
| chr11_+_77526 | 4.48 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr12_-_212843 | 4.48 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr6_-_35446110 | 4.47 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr12_+_18606140 | 4.46 |
ENSDART00000161128
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr18_+_41495841 | 4.45 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr25_+_20119466 | 4.40 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr22_+_4707663 | 4.29 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr20_-_45661049 | 4.29 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr6_+_42819337 | 4.28 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr3_-_28075756 | 4.21 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr11_-_24191928 | 4.12 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr4_+_19534833 | 4.08 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr3_-_46818001 | 4.05 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr5_-_2689753 | 4.05 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr2_-_7431590 | 4.03 |
ENSDART00000185699
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr7_-_38612230 | 4.00 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr8_+_23165749 | 3.97 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr2_-_31936966 | 3.97 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr16_-_12173554 | 3.96 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr1_-_20271138 | 3.92 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr7_-_26408472 | 3.83 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr16_+_68069 | 3.81 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr9_+_17348745 | 3.81 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr13_+_25428677 | 3.66 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr14_+_36220479 | 3.66 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr6_+_40661703 | 3.64 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr24_+_25069609 | 3.58 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr7_-_52849913 | 3.55 |
ENSDART00000174133
ENSDART00000172951 |
map1aa
|
microtubule-associated protein 1Aa |
| chr4_-_9173552 | 3.54 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr5_+_31283576 | 3.54 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr19_-_25113660 | 3.51 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_-_19395476 | 3.50 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_+_26340736 | 3.47 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr25_-_8602437 | 3.45 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr14_+_15597049 | 3.45 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr14_+_15543331 | 3.44 |
ENSDART00000167025
|
si:dkey-203a12.7
|
si:dkey-203a12.7 |
| chr25_+_37348730 | 3.42 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr5_-_40510397 | 3.42 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr6_+_52263236 | 3.38 |
ENSDART00000144174
|
esyt1b
|
extended synaptotagmin-like protein 1b |
| chr25_-_32888115 | 3.38 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr2_+_38924975 | 3.37 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr13_+_23214100 | 3.36 |
ENSDART00000163393
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_-_46273938 | 3.35 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr22_-_18746508 | 3.32 |
ENSDART00000003929
|
midn
|
midnolin |
| chr11_-_43137712 | 3.29 |
ENSDART00000173089
|
si:zfos-1837d3.1
|
si:zfos-1837d3.1 |
| chr1_+_1599979 | 3.24 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr17_-_14726824 | 3.24 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr25_-_29134654 | 3.22 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr13_+_36633355 | 3.21 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr12_+_11080776 | 3.20 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr5_-_14390445 | 3.17 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr10_+_15777064 | 3.16 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr14_+_15495088 | 3.16 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr8_+_28900689 | 3.15 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr3_+_35005730 | 3.14 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr19_+_14573998 | 3.14 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr20_+_52554352 | 3.14 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr17_-_42213285 | 3.02 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr7_+_7048245 | 3.01 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr3_-_46817838 | 2.99 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr7_+_31051213 | 2.99 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr18_-_39702327 | 2.95 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr23_+_9057999 | 2.94 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr2_-_17828279 | 2.92 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr1_-_59176949 | 2.91 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr25_-_23526058 | 2.91 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr3_-_6767440 | 2.91 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr18_-_12327426 | 2.91 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr4_+_2619132 | 2.87 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr23_+_44307996 | 2.85 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr17_-_20897407 | 2.84 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr24_+_31334209 | 2.83 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr5_+_72377851 | 2.83 |
ENSDART00000160479
|
lman2la
|
lectin, mannose-binding 2-like a |
| chr22_-_12693833 | 2.82 |
ENSDART00000129768
ENSDART00000044574 |
adarb1a
|
adenosine deaminase, RNA-specific, B1a |
| chr13_-_39947335 | 2.82 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr6_+_42818963 | 2.81 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr1_+_604127 | 2.79 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr1_+_32528097 | 2.77 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr5_+_32791245 | 2.76 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr8_+_34731982 | 2.75 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr17_+_12698532 | 2.75 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr11_+_6819050 | 2.74 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr8_+_8298439 | 2.73 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr5_-_42180205 | 2.73 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr2_+_8112449 | 2.73 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr1_-_7659870 | 2.73 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr23_+_41799748 | 2.72 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr12_+_35650321 | 2.70 |
ENSDART00000190446
|
BX255898.1
|
|
| chr10_+_26571174 | 2.68 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr5_+_65492183 | 2.66 |
ENSDART00000162804
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr2_+_47582488 | 2.60 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr12_-_13886952 | 2.60 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr11_-_11625369 | 2.59 |
ENSDART00000112328
|
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr3_-_30685401 | 2.57 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr22_+_1291651 | 2.55 |
ENSDART00000159296
|
si:ch73-138e16.4
|
si:ch73-138e16.4 |
| chr20_+_27298783 | 2.55 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr25_+_27493444 | 2.52 |
ENSDART00000112299
|
gpr37a
|
G protein-coupled receptor 37a |
| chr21_+_15312542 | 2.51 |
ENSDART00000144308
|
si:dkey-11o15.5
|
si:dkey-11o15.5 |
| chr12_+_19036380 | 2.51 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr13_+_12739283 | 2.51 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr3_+_62217479 | 2.51 |
ENSDART00000175012
|
BX470259.4
|
|
| chr21_+_7900107 | 2.50 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr3_-_39488639 | 2.48 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr25_-_18125769 | 2.45 |
ENSDART00000140484
|
kitlga
|
kit ligand a |
| chr10_+_38643304 | 2.44 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr5_+_483965 | 2.44 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr25_-_7999756 | 2.40 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr23_-_2081554 | 2.38 |
ENSDART00000179805
|
ndnf
|
neuron-derived neurotrophic factor |
| chr3_-_39488482 | 2.37 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr1_+_47459723 | 2.37 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr24_+_2801765 | 2.34 |
ENSDART00000172385
|
LYRM4
|
si:ch211-152c8.4 |
| chr2_+_59015878 | 2.32 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr8_+_24854600 | 2.32 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr19_+_2602903 | 2.32 |
ENSDART00000033132
|
fam126a
|
family with sequence similarity 126, member A |
| chr19_-_11031145 | 2.31 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr11_-_11625630 | 2.27 |
ENSDART00000161821
ENSDART00000193152 |
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr15_+_46344655 | 2.21 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr6_-_18976168 | 2.20 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr9_-_24046287 | 2.20 |
ENSDART00000184313
|
ackr3a
|
atypical chemokine receptor 3a |
| chr8_+_22289320 | 2.19 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr24_-_41478917 | 2.19 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr17_+_30587333 | 2.17 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr5_-_23517747 | 2.17 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| chr15_-_23342752 | 2.16 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr3_+_24537023 | 2.16 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr14_+_12316581 | 2.15 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr18_+_18612388 | 2.13 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr25_+_24250247 | 2.12 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr1_+_31942961 | 2.09 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr16_-_12316979 | 2.08 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr19_+_1964005 | 2.07 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr14_+_15331486 | 2.06 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr15_-_5467477 | 2.04 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr16_-_12173399 | 2.03 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr22_-_3564563 | 2.01 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr25_-_10564721 | 1.99 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr16_+_11151699 | 1.97 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr5_-_24270989 | 1.96 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr23_+_16633951 | 1.95 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr5_+_31168096 | 1.95 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr5_-_64431927 | 1.93 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr3_+_16976095 | 1.92 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr12_+_23762966 | 1.88 |
ENSDART00000152942
ENSDART00000181725 |
jcada
|
junctional cadherin 5 associated a |
| chr20_-_54564018 | 1.87 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr12_-_4346085 | 1.85 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr10_+_42542517 | 1.85 |
ENSDART00000005496
|
kctd9b
|
potassium channel tetramerization domain containing 9b |
| chr16_-_22775480 | 1.85 |
ENSDART00000141778
ENSDART00000145585 ENSDART00000125963 ENSDART00000127570 |
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr14_-_21218891 | 1.83 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr4_-_58846245 | 1.81 |
ENSDART00000170777
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr3_-_27915270 | 1.81 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr24_+_19542323 | 1.81 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr10_+_1668106 | 1.81 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr10_+_22891126 | 1.80 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr3_+_40809011 | 1.80 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr2a+egr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 1.9 | 5.6 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 1.4 | 4.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.3 | 4.0 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 1.3 | 6.4 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 1.2 | 4.9 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.1 | 3.3 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 1.1 | 4.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.0 | 2.9 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 1.0 | 3.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.9 | 2.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.9 | 2.8 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.9 | 6.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.9 | 2.7 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.9 | 5.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.9 | 5.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.8 | 2.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.8 | 4.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.7 | 3.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.7 | 3.5 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.7 | 13.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.7 | 6.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.6 | 2.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.6 | 2.4 | GO:0021856 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.6 | 5.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 3.2 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.5 | 2.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 5.7 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.5 | 1.9 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.5 | 2.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 3.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.4 | 3.0 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 1.6 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 5.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 2.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 3.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.4 | 1.5 | GO:0071548 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.4 | 3.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 5.0 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.3 | 2.4 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.3 | 1.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 8.9 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.3 | 0.6 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.3 | 3.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 1.2 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 4.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 6.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 7.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 5.9 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.3 | 3.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 1.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 1.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.3 | 3.9 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 1.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 3.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 1.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.2 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 2.7 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 1.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 8.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.2 | 6.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 2.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 1.8 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 3.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 1.6 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.2 | 3.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.5 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.2 | 3.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 0.8 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.2 | 6.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 4.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 1.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.2 | 1.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 2.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.0 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 4.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.2 | 2.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 2.9 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 2.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 7.1 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 2.4 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.1 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 2.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 1.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 7.4 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 1.0 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 3.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.0 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.4 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 4.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.3 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 2.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 8.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 2.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.0 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 11.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 3.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 4.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 6.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 1.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 3.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 2.2 | GO:0048883 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 2.0 | GO:1901184 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) regulation of ERBB signaling pathway(GO:1901184) |
| 0.1 | 2.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.9 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 2.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 2.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 2.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.8 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 2.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 3.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.4 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.1 | 0.4 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.5 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 4.4 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 6.3 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 7.5 | GO:0071772 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 4.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 5.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 1.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 10.6 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 2.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 3.0 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 1.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.6 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.3 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 1.2 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.0 | GO:0003139 | heart field specification(GO:0003128) secondary heart field specification(GO:0003139) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.1 | 4.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.9 | 6.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.7 | 2.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 2.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.6 | 10.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.6 | 6.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 5.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.5 | 3.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 1.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 12.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.3 | 1.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 1.6 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 13.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 7.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 1.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 3.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 7.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 12.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 2.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 3.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 8.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 6.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 4.4 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 12.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.8 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 5.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 1.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.9 | GO:0031256 | leading edge membrane(GO:0031256) ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 24.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 13.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.7 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 3.6 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.3 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.0 | 9.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.9 | 2.8 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.9 | 2.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.8 | 4.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.7 | 4.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 3.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.7 | 10.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 3.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 2.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.6 | 2.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 7.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 2.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 2.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.5 | 4.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 5.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 3.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 9.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.5 | 1.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 7.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.5 | 7.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 5.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.5 | 3.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 3.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.4 | 1.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 3.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 3.8 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 1.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 2.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.3 | 9.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 4.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 1.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 2.6 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.3 | 1.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 3.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 1.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 1.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 4.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 3.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 1.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 1.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.3 | 1.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 2.7 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.2 | 2.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 2.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 10.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 7.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 1.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 4.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 4.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 5.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 1.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 6.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 3.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.8 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 0.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 2.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 6.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 3.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 4.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 5.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 13.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 3.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 2.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 6.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 11.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 5.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 2.4 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 3.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 6.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 7.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.8 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 11.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 2.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 11.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 3.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 4.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 6.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 5.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 5.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 5.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 7.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 7.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.6 | 15.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 6.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 5.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 8.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.3 | 3.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 7.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 6.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |