Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
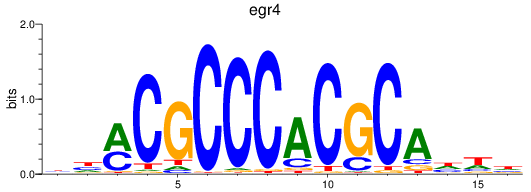
Results for egr4
Z-value: 0.34
Transcription factors associated with egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr4
|
ENSDARG00000077799 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr4 | dr11_v1_chr23_+_45579497_45579497 | 0.40 | 5.8e-05 | Click! |
Activity profile of egr4 motif
Sorted Z-values of egr4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_48753873 | 4.81 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr10_+_19554604 | 2.83 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr24_-_24146875 | 2.71 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr12_+_35650321 | 2.49 |
ENSDART00000190446
|
BX255898.1
|
|
| chr2_-_31301929 | 2.39 |
ENSDART00000191992
ENSDART00000190723 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr7_-_44963154 | 2.37 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr5_+_64739762 | 2.14 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr8_+_23165749 | 2.01 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr2_+_38924975 | 1.96 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr23_+_41799748 | 1.61 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr19_+_342094 | 1.47 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr3_+_31897029 | 1.37 |
ENSDART00000146275
|
strada
|
ste20-related kinase adaptor alpha |
| chr14_+_36220479 | 1.31 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr14_-_35400671 | 1.28 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr11_+_12719944 | 1.25 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_-_15657029 | 1.21 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr23_+_41800052 | 1.20 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr3_+_24537023 | 1.16 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr6_+_40661703 | 1.13 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr18_+_3169579 | 1.12 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_+_12720171 | 1.09 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr25_-_207214 | 1.06 |
ENSDART00000193448
|
FP236318.3
|
|
| chr5_-_14390445 | 1.03 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr23_+_40460333 | 1.00 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr13_+_35925490 | 0.98 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr6_+_42819337 | 0.95 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr16_+_27614989 | 0.88 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr5_-_2689753 | 0.88 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr17_+_15297398 | 0.87 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr10_-_15919839 | 0.84 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr14_-_21218891 | 0.83 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr25_+_13406069 | 0.83 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr2_+_16710889 | 0.80 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr12_-_7639120 | 0.78 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr1_-_7917062 | 0.73 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr6_-_23117348 | 0.71 |
ENSDART00000154941
ENSDART00000154252 |
ten1
|
TEN1 CST complex subunit |
| chr3_-_32180796 | 0.71 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr1_-_10473630 | 0.68 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr20_-_10065130 | 0.65 |
ENSDART00000134439
|
znf770
|
zinc finger protein 770 |
| chr6_+_2271559 | 0.65 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr24_+_24831727 | 0.64 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr22_+_1806235 | 0.62 |
ENSDART00000158966
|
znf1168
|
zinc finger protein 1168 |
| chr20_+_28803642 | 0.60 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr16_-_40508624 | 0.58 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr21_-_43398457 | 0.55 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr12_-_23129453 | 0.53 |
ENSDART00000077453
|
armc4
|
armadillo repeat containing 4 |
| chr5_+_51443009 | 0.52 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr20_+_9211237 | 0.52 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr21_-_43398122 | 0.44 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr6_+_42818963 | 0.44 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr18_+_20481982 | 0.43 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr23_+_9067131 | 0.43 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr22_+_10713713 | 0.43 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr3_-_1520657 | 0.41 |
ENSDART00000053201
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr5_+_61941610 | 0.40 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr9_-_30363770 | 0.39 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr13_-_44834895 | 0.39 |
ENSDART00000135573
|
si:dkeyp-2e4.5
|
si:dkeyp-2e4.5 |
| chr13_-_49666615 | 0.38 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr3_+_22984098 | 0.37 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr24_-_39354829 | 0.37 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr16_+_53519048 | 0.36 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr22_-_20720427 | 0.33 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr22_-_9839003 | 0.28 |
ENSDART00000193573
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr11_+_14049131 | 0.28 |
ENSDART00000172471
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr3_-_7106678 | 0.27 |
ENSDART00000175293
ENSDART00000187732 ENSDART00000158652 |
zgc:113102
BX005085.2
|
zgc:113102 |
| chr23_-_524780 | 0.27 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr5_-_21030934 | 0.27 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr21_-_31238244 | 0.25 |
ENSDART00000159678
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr8_+_45294767 | 0.24 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr4_-_9579299 | 0.16 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr22_-_34872533 | 0.16 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr3_+_16722014 | 0.15 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr12_+_499881 | 0.15 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr11_+_3308656 | 0.15 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr21_+_33172526 | 0.14 |
ENSDART00000183532
|
arl3l1
|
ADP-ribosylation factor-like 3, like 1 |
| chr2_+_51194657 | 0.14 |
ENSDART00000161381
|
lrrc24
|
leucine rich repeat containing 24 |
| chr1_-_47431453 | 0.13 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr4_+_70556298 | 0.09 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr18_+_8901846 | 0.08 |
ENSDART00000132109
ENSDART00000144247 |
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr7_+_41340520 | 0.06 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr8_-_52859301 | 0.04 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr16_-_54405976 | 0.03 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr3_+_16662209 | 0.03 |
ENSDART00000190556
|
zgc:55558
|
zgc:55558 |
| chr23_+_804119 | 0.02 |
ENSDART00000148163
|
frmd4bb
|
FERM domain containing 4Bb |
| chr19_+_5480327 | 0.01 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr18_+_20482369 | 0.00 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr13_-_36418921 | 0.00 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 1.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 2.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 1.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.7 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.2 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.9 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 4.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 1.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.0 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0030534 | adult behavior(GO:0030534) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 2.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 2.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.3 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.2 | 2.8 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 4.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 1.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 2.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 1.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |